













| General Information: |
|
| Name(s) found: |
PUF3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 879 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMNMDMDMD MELASIVSSL SALSHSNNNG GQAAAAGIVN GGAAGSQQIG GFRRSSFTTA 60
61 NEVDSEILLL HGSSESSPIF KKTALSVGTA PPFSTNSKKF FGNGGNYYQY RSTDTASLSS 120
121 ASYNNYHTHH TAANLGKNNK VNHLLGQYSA SIAGPVYYNG NDNNNSGGEG FFEKFGKSLI 180
181 DGTRELESQD RPDAVNTQSQ FISKSVSNAS LDTQNTFEQN VESDKNFNKL NRNTTNSGSL 240
241 YHSSSNSGSS ASLESENAHY PKRNIWNVAN TPVFRPSNNP AAVGATNVAL PNQQDGPANN 300
301 NFPPYMNGFP PNQFHQGPHY QNFPNYLIGS PSNFISQMIS VQIPANEDTE DSNGKKKKKA 360
361 NRPSSVSSPS SPPNNSPFPF AYPNPMMFMP PPPLSAPQQQ QQQQQQQQQE DQQQQQQQEN 420
421 PYIYYPTPNP IPVKMPKDEK TFKKRNNKNH PANNSNNANK QANPYLENSI PTKNTSKKNA 480
481 SSKSNESTAN NHKSHSHSHP HSQSLQQQQQ TYHRSPLLEQ LRNSSSDKNS NSNMSLKDIF 540
541 GHSLEFCKDQ HGSRFIQREL ATSPASEKEV IFNEIRDDAI ELSNDVFGNY VIQKFFEFGS 600
601 KIQKNTLVDQ FKGNMKQLSL QMYACRVIQK ALEYIDSNQR IELVLELSDS VLQMIKDQNG 660
661 NHVIQKAIET IPIEKLPFIL SSLTGHIYHL STHSYGCRVI QRLLEFGSSE DQESILNELK 720
721 DFIPYLIQDQ YGNYVIQYVL QQDQFTNKEM VDIKQEIIET VANNVVEYSK HKFASNVVEK 780
781 SILYGSKNQK DLIISKILPR DKNHALNLED DSPMILMIKD QFANYVIQKL VNVSEGEGKK 840
841 LIVIAIRAYL DKLNKSNSLG NRHLASVEKL AALVENAEV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
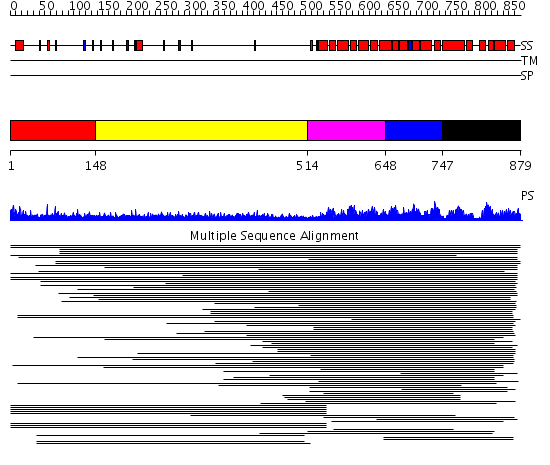
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 3.404997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [148..513] | 14.59 | RBP1 | |
| 3 | View Details | [514..647] | 973.9794 | Pumilio 1 | |
| 4 | View Details | [648..746] | 973.9794 | Pumilio 1 | |
| 5 | View Details | [747..879] | 973.9794 | Pumilio 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)