













| General Information: |
|
| Name(s) found: |
NUP2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 720 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRVADAQI QRETYDSNES DDDVTPSTKV ASSAVMNRRK IAMPKRRMAF KPFGSAKSDE 60
61 TKQASSFSFL NRADGTGEAQ VDNSPTTESN SRLKALNLQF KAKVDDLVLG KPLADLRPLF 120
121 TRYELYIKNI LEAPVKSIEN PTQTKGNDAK PAKVEDVQKS SDSSSEDEVK VEGPKFTIDA 180
181 KPPISDSVFS FGPKKENRKK DESDSENDIE IKGPEFKFSG TVSSDVFKLN PSTDKNEKKT 240
241 ETNAKPFSFS SATSTTEQTK SKNPLSLTEA TKTNVDNNSK AEASFTFGTK HAADSQNNKP 300
301 SFVFGQAAAK PSLEKSSFTF GSTTIEKKND ENSTSNSKPE KSSDSNDSNP SFSFSIPSKN 360
361 TPDASKPSFS FGVPNSSKNE TSKPVFSFGA ATPSAKEASQ EDDNNNVEKP SSKPAFNLIS 420
421 NAGTEKEKES KKDSKPAFSF GISNGSESKD SDKPSLPSAV DGENDKKEAT KPAFSFGINT 480
481 NTTKTADTKA PTFTFGSSAL ADNKEDVKKP FSFGTSQPNN TPSFSFGKTT ANLPANSSTS 540
541 PAPSIPSTGF KFSLPFEQKG SQTTTNDSKE ESTTEATGNE SQDATKVDAT PEESKPINLQ 600
601 NGEEDEVALF SQKAKLMTFN AETKSYDSRG VGEMKLLKKK DDPSKVRLLC RSDGMGNVLL 660
661 NATVVDSFKY EPLAPGNDNL IKAPTVAADG KLVTYIVKFK QKEEGRSFTK AIEDAKKEMK 720
721 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
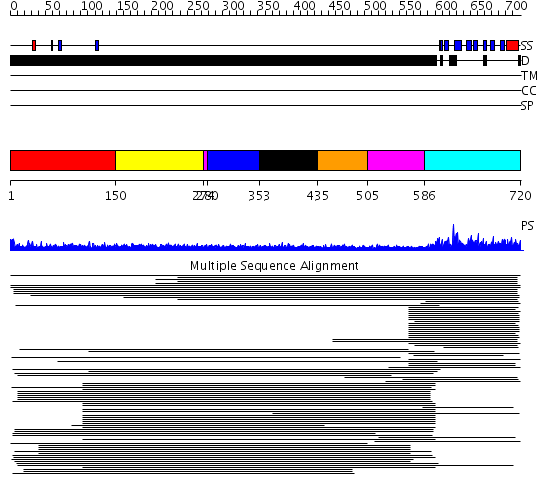
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [150..273] | 14.0 | Sec24 | |
| 3 | View Details | [274..279] [505..585] |
11.01 | No description for 1l8mA was found. | |
| 4 | View Details | [280..352] | 11.01 | No description for 1l8mA was found. | |
| 5 | View Details | [353..434] | 11.01 | No description for 1l8mA was found. | |
| 6 | View Details | [435..504] | 11.01 | No description for 1l8mA was found. | |
| 7 | View Details | [586..720] | 134.190949 | Nuclear pore complex protein Nup358 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)