













| General Information: |
|
| Name(s) found: |
MMS4_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQIVDFVED KDSRNDASIQ IIDGPSNVEI IALSESMDQD ECKRAHVSSA EMIPSSPQRK 60
61 SVSNDVENVD LNKSIELSAP FFQDISISKL DDFSTTVNSI IDSSLRNENN AKGNAKKLLD 120
121 DLISDEWSAD LESSGKKHNK SQYNLRDIAE KWGVQSLKNP EPIAVDCEYK TQGIGKTNSD 180
181 ISDSPKSQIG AADILFDFPL SPVKHENPTE EKHNSIANEN SSPDNSLKPA GKQNHGEDGT 240
241 SMAKRVYNKG EDEQEHLPKG KKRTIALSRT LINSTKLPDT VELNLSKFLD SSDSITTDVL 300
301 STPAKGSNIV RTGSQPIFSN ANCFQEAKRS KTLTAEDPKC TKNTAREVSQ LENYIAYGQY 360
361 YTREDSKNKI RHLLKENKNA FKRVNQIYRD NIKARSQMII EFSPSLLQLF KKGDSDLQQQ 420
421 LAPAVVQSSY NDSMPLLRFL RKCDSIYDFS NDFYYPCDPK IVEENVLILY YDAQEFFEQY 480
481 TSQKKELYRK IRFFSKNGKH VILILSDINK LKRAIFQLEN EKYKARVEQR LSGTEEALRP 540
541 RSKKSSQVGK LGIKKFDLEQ RLRFIDREWH VKIHTVNSHM EFINSLPNLV SLIGKQRMDP 600
601 AIRYMKYAHL NVKSAQDSTE TLKKTFHQIG RMPEMKANNV VSLYPSFQSL LEDIEKGRLQ 660
661 SDNEGKYLMT EAVEKRLYKL FTCTDPNDTI E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
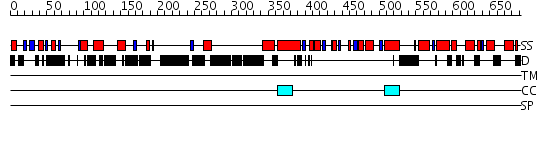
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..394] | 1.28 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | |
| 2 | View Details | [395..456] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [457..691] | 1.6 | XPF from Aeropyrum pernix, complex with DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)