













| General Information: |
|
| Name(s) found: |
IQG1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1495 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAYSGSPSK PGNNNSYLNR YVENLGTNVT PPLRPQSSSK INSSLNIASP SHLKTKTSAS 60
61 NSSATILSKK VESSVSKLKP SLPNKLVGKY TVDLSNYSKI ELRYYEFLCR VSEVKIWIEA 120
121 VIEEALPSEI ELCVGDSLRN GVFLAKLTQR INPDLTTVIF PAGDKLQFKH TQNINAFFGL 180
181 VEHVGVPDSF RFELQDLYNK KNIPQVFETL HILISMINKK WPGKTPALTN VSGQISFTKE 240
241 EIAACKKAWP RIRDFKSLGT NINTAPASPE EPKEKRSGLI KDFNKFERPN IPVEEILITP 300
301 RKNITDANCS DFSNTPSPYN EAPKMSNLDV VVEKRKFTPI EPSLLGPTPS LEYSPIKNKS 360
361 LSYYSPTISK YLTYDTEFYT RRSRAREEDL NYYQTFKYSP SHYSPMRRER MTEEQFLEKV 420
421 VQLQNICRGV NTRFNLYIQK RLLNLFEQDI LRFQACLRGN KFRVLSSMYL PIRRAKIDVP 480
481 HVEAIQSRIK GSRIRYKYDK LKFTLSRFSC TVELLQAYCR SKLLKTTVNT KLNDIEISHY 540
541 PLTKLQSYMR ASYVRKKVMS LNTKLNDERE SIMKFSAIIR GNVVRCSEDA ILSAVHDVHK 600
601 ENISKLQSLI RGIFTRSCLA SIIYSLGKEN CNIIQLSACI RGNAVRHKVQ SLFAPENNLS 660
661 ETVHDLQGLV RGILVRYTLD LVDDIVEYNN LALFQAFSRG ALVRESLDQK SSFYKRNVRS 720
721 VIMIQSWIRK SLQRSAYLEL LDCPNPSLWA VKKFVHLLNG TATIEEVQNQ LESCQASLDS 780
781 ENMKKERLLK SIRQQLNING VLDKFGLLKD KDHELGISDS TIPKSKYQKY EKLFYMLQVD 840
841 PSYWKLLYLK EPEFVAKNVY MTFGTVNQRM NDRERSYFTR FVCEMLQNAI NEAPSIESFL 900
901 DNRSQFWQTI LQDFLRRESP EFFSIIVPVL DYLSDPVVDF ESDPYKIYQE IHGFSSPQHC 960
961 SPVDDASTKN KFIDNLRCLW HAIEMVAEIY TRKVHTIPVE IRYLCTKIFC YAADKNIEEI1020
1021 DSLRAISSIL VNVFVSEYLV NREYYGYKDS NVQKNNQKID ILMKSLATVF EIKNFDGFLD1080
1081 PLNQYANEIK PHIKDVLYNV LVDPEYEQEG DRLIYLDMVS PSPKLELLTE KVLEISGKFE1140
1141 EYLNEFPEAD ILHDILEKNL DNSSFPRSGR VTLELDASAY RFLVSDDKMR KIYDQVKRAF1200
1201 VYMMQIEDVD TNLYDLSIST ILPQDEPNFA NFLEQNPKIR DDPMIQKLKP LKYFTLKNVT1260
1261 LKKIHELEST GTFCSSDNKL QNFLNDIANT IKNPNYAIDY VTQEIYITKE TLTKISEMNH1320
1321 SLDIELSRLK KHVDHTIKDF QKAKDFSPVH KSKFGNFKNA VKKVQGRERS ELQGMKFKWN1380
1381 TKQLYERGVL KTIRGEKLAE LTVKVFGSSG PKFPDIIFKI STSDGSRFGI QMIDKRKGPD1440
1441 KRYSDDVDSF SFKDLIKTQV EPKIETWKLF HSNVVVNNSQ LLHLIVSFFY KRNAL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
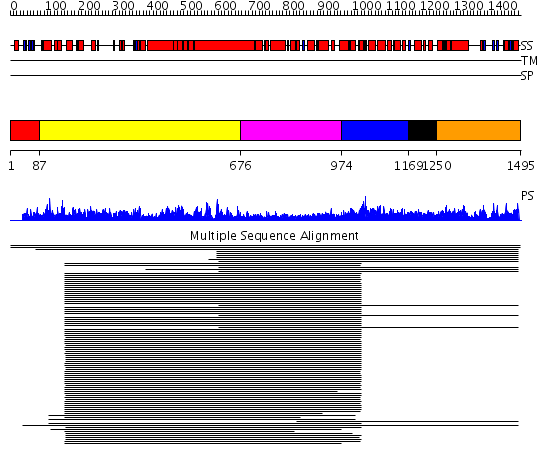
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..675] | 149.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [676..973] | 149.0 | Heavy meromyosin subfragment | |
| 4 | View Details | [974..1168] | 57.0 | GAP related domain of neurofibromin | |
| 5 | View Details | [1169..1249] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1250..1495] | 2.024962 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)