













| General Information: |
|
| Name(s) found: |
DPOA2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGSIDVITH FGPDADKPEI ITALENLTKL HALSVEDLYI KWEQFSNQRR QTHTDLTSKN 60
61 IDEFKQFLQL QMEKRANQIS SSSKVNTSTK KPVIKKSLNS SPLFGLSIPK TPTLKKRKLH 120
121 GPFSLSDSKQ TYNVGSEAET NEKGNSSLKL EFTPGMAEDA VGDSAPLSHA KSSDAKTPGS 180
181 STFQTPTTNT PTTSRQNVPA GEILDSLNPE NIEISSGNPN VGLLSTEEPS YNQVKVEPFY 240
241 DAKKYKFRTM RQNLQEASDV LDDQIESFTK IIQNHYKLSP NDFADPTIQS QSEIYAVGRI 300
301 VPDSPTYDKF LNPESLSLET SRMGGVGRRV RLDLSQVNEL SFFLGQIVAF KGKNANGDYF 360
361 TVNSILPLPY PNSPVSTSQE LQEFQANLEG SSLKVIVTCG PYFANDNFSL ELLQEFIDSI 420
421 NNEVKPHVLI MFGPFIDITH PLIASGKLPN FPQFKTQPKT LDELFLKLFT PILKTISPHI 480
481 QTVLIPSTKD AISNHAAYPQ ASLIRKALQL PKRNFKCMAN PSSFQINEIY FGCSNVDTFK 540
541 DLKEVIKGGT TSSRYRLDRV SEHILQQRRY YPIFPGSIRT RIKPKDVSTK KETNDMESKE 600
601 EKVYEHISGA DLDVSYLGLT EFVGGFSPDI MIIPSELQHF ARVVQNVVVI NPGRFIRATG 660
661 NRGSYAQITV QCPDLEDGKL TLVEGEEPVY LHNVWKRARV DLIAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
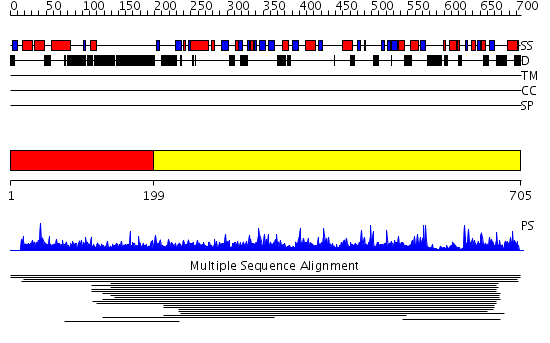
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..198] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [199..705] | 3.0129 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)