













| General Information: |
|
| Name(s) found: |
BPL1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 690 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNVLVYNGPG TTPGSVKHAV ESLRDFLEPY YAVSTVNVKV LQTEPWMSKT SAVVFPGGAD 60
61 LPYVQACQPI ISRLKHFVSK QGGVFIGFCA GGYFGTSRVE FAQGDPTMEV SGSRDLRFFP 120
121 GTSRGPAYNG FQYNSEAGAR AVKLNLPDGS QFSTYFNGGA VFVDADKFDN VEILATYAEH 180
181 PDVPSSDSGK GQSENPAAVV LCTVGRGKVL LTGPHPEFNV RFMRKSTDKH FLETVVENLK 240
241 AQEIMRLKFM RTVLTKTGLN CNNDFNYVRA PNLTPLFMAS APNKRNYLQE MENNLAHHGM 300
301 HANNVELCSE LNAETDSFQF YRGYRASYDA ASSSLLHKEP DEVPKTVIFP GVDEDIPPFQ 360
361 YTPNFDMKEY FKYLNVQNTI GSLLLYGEVV TSTSTILNNN KSLLSSIPES TLLHVGTIQV 420
421 SGRGRGGNTW INPKGVCAST AVVTMPLQSP VTNRNISVVF VQYLSMLAYC KAILSYAPGF 480
481 SDIPVRIKWP NDLYALSPTY YKRKNLKLVN TGFEHTKLPL GDIEPAYLKI SGLLVNTHFI 540
541 NNKYCLLLGC GINLTSDGPT TSLQTWIDIL NEERQQLHLD LLPAIKAEKL QALYMNNLEV 600
601 ILKQFINYGA AEILPSYYEL WLHSNQIVTL PDHGNTQAMI TGITEDYGLL IAKELVSGSS 660
661 TQFTGNVYNL QPDGNTFDIF KSLIAKKVQS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
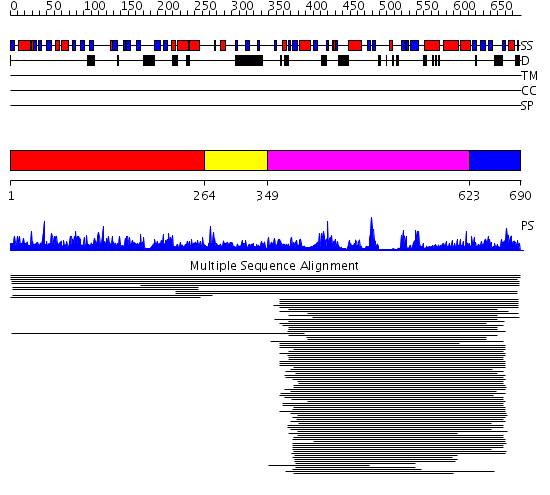
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..263] | 4.010916 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [264..348] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [349..622] | 85.68867 | Biotin repressor, N-terminal domain; Biotin repressor/biotin holoenzyme synthetase, C-terminal domain; Biotin repressor/biotin holoenzyme synthetase, catalytic (central) domain | |
| 4 | View Details | [623..690] | 85.68867 | Biotin repressor, N-terminal domain; Biotin repressor/biotin holoenzyme synthetase, C-terminal domain; Biotin repressor/biotin holoenzyme synthetase, catalytic (central) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)