













| General Information: |
|
| Name(s) found: |
BNR1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1375 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSSPNKKTY RYPRRSLSLH ARDRVSEARK LEELNLNDGL VAAGLQLVGV ALEKQGTGSH 60
61 IYMKQKNFSA NDVSSSPMVS EEVNGSEMDF NPKCMPQDAS LVERMFDELL KDGTFFWGAA 120
121 YKNLQNISLR RKWLLICKIR SSNHWGKKKV TSSTTYSTHL ATNELAENAH FLDGLVRNLS 180
181 TGGMKLSKAL YKLEKFLRKQ SFLQLFLKDE IYLTTLIEKT LPLISKELQF VYLRCFKILM 240
241 NNPLARIRAL HSEPLIRWFT ELLTDQNSNL KCQLLSMELL LLLTYVEGST GCELIWDQLS 300
301 ILFTDWLEWF DKILADDIAI HSSLYLNWNQ LKIDYSTTFL LLINSILQGF NNKTALEILN 360
361 FLKKNNIHNT ITFLELAYKD DPNSVVIMEQ IKQFKSKESA IFDSMIKTTN DTNSLHPTKD 420
421 IARIESEPLC LENCLLLKAK DSPVEAPINE IIQSLWKILD SQKPYSESIK LLKLINSLLF 480
481 YLIDSFQVST NPSFDETLES AENVDYVFQD SVNKLLDSLQ SDEIARRAVT EIDDLNAKIS 540
541 HLNEKLNLVE NHDKDHLIAK LDESESLISL KTKEIENLKL QLKATKKRLD QITTHQRLYD 600
601 QPPSLASSNL SIAGSIIKNN SHGNIIFQNL AKKQQQQQKI SLPKRSTSLL KSKRVTSLSS 660
661 YLTDANNENE SQNESEDKSK DSLFQRSTST INFNIPSMKN ITNMQNVSLN SILSELEFSN 720
721 SLGTQPNYQS SPVLSSVSSS PKLFPRLSSD SLDNGIQLVP EVVKLPQLPP PPPPPPPPPL 780
781 PQSLLTEAEA KPDGVSCIAA PAPPPLPDLF KTKTCGAVPP PPPPPPLPES LSMNKGPSNH 840
841 DLVTPPAPPL PNGLLSSSSV SINPTTTDLK PPPTEKRLKQ IHWDKVEDIK DTLWEDTFQR 900
901 QETIKELQTD GIFSQIEDIF KMKSPTKIAN KRNAESSIAL SSNNGKSSNE LKKISFLSRD 960
961 LAQQFGINLH MFSQLSDMEF VMKVLNCDND IVQNVNILKF FCKEELVNIP KSMLNKYEPY1020
1021 SQGKDGKAVS DLQRADRIFL ELCINLRFYW NARSKSLLTL STYERDYYDL IFKLQKIDDA1080
1081 ISHLNRSPKF KSLMFIITEI GNHMNKRIVK GIKLKSLTKL AFVRSSIDQN VSFLHFIEKV1140
1141 IRIKYPDIYG FVDDLKNIED LGKISLEHVE SECHEFHKKI EDLVTQFQVG KLSKEENLDP1200
1201 RDQIIKKVKF KINRAKTKSE LLIGQCKLTL IDLNKLMKYY GEDPKDKESK NEFFQPFIEF1260
1261 LAMFKKCAKE NIEKEEMERV YEQRKSLLDM RTSSNKKSNG SDENDGEKVN RDAVDLLISK1320
1321 LREVKKDPEP LRRRKSTKLN EIAINVHEGD VKTRKDEDHV LLERTHAMLN DIQNI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
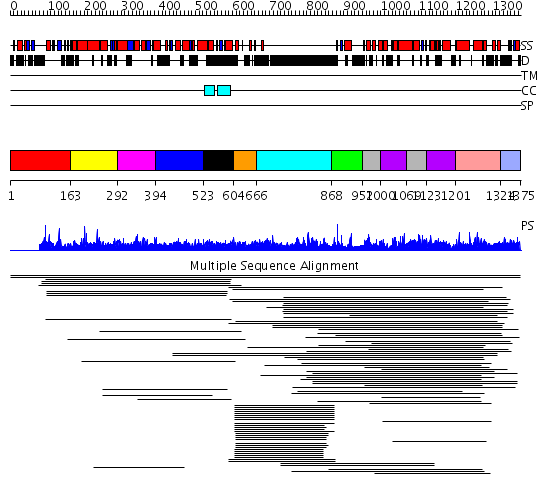
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | 1.009996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [163..291] | 8.79 | beta-Catenin | |
| 3 | View Details | [292..393] | 8.79 | beta-Catenin | |
| 4 | View Details | [394..522] | 8.79 | beta-Catenin | |
| 5 | View Details | [523..603] | 8.79 | beta-Catenin | |
| 6 | View Details | [604..665] | 6.154902 | Dynamin | |
| 7 | View Details | [666..867] | 11.94 | Fibrinogen | |
| 8 | View Details | [868..951] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [952..999] [1069..1122] |
7.79 | DNA-directed RNA polymerase alpha(2), beta, beta-prime and omega subunits | |
| 10 | View Details | [1000..1068] [1123..1200] |
7.79 | DNA-directed RNA polymerase alpha(2), beta, beta-prime and omega subunits | |
| 11 | View Details | [1201..1323] | 7.79 | DNA-directed RNA polymerase alpha(2), beta, beta-prime and omega subunits | |
| 12 | View Details | [1324..1375] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 12 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)