













| General Information: |
|
| Name(s) found: |
BCH2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 765 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFLWGSTKS KKGKNKKAAG SLPSGVVPQQ RVKPTRKNVP IDYPRTLEKV HGESLIFRTS 60
61 LLSELVSTGK SGIGPPDLIH CTELDKFHDE KIGEFFYITG IDASSVSMPI AFLKLIKWND 120
121 GKKLKSASLK NDDITTYCTF NIFQKLDIRL RYESEDVYQV NIVDCLNGNN EIPLSDLIWE 180
181 ETFVSCCIRS VIINSDFERK IPGLVELPFV FENRCASDYK RVIDSLCKFL PRFLECGWDS 240
241 TKSVYATILN NYLTESLLVF LSITPEFITD YAIQVLDNLM TNDPSNSRYY AIVIISIMER 300
301 SNDRDVEMIK RIHEILDLLL PVLYGLPSDE PYISDLINCI TDVLSIQARF LLNNNDYELS 360
361 LSISTLATNL SSDNFESWYL LSKGYIFSQQ YDKALLSINS MPCLAEYDIV KQAQINAFKF 420
421 YMNYYKAPLC HSREHCTMTS HELNHLMNIM HYENELELKT IIFGRTVMPN ESKYGCIEEI 480
481 WNKSCLELGP ICGPQSDNLI NFVSQQEVNT VGDMLLLKRS KETRQESWFI KQVRLLLMEL 540
541 VARIGWNALL QLRSDVFVME SKFKMIESSD KLSTELRQKR LCQRWFDAMF LDVYEDLSIS 600
601 TSSQENKATA KYSGLEWELL GLTLLRVSDL PDAVACLRTS ILARFDPISC HHLLNFYLTM 660
661 DFNDEFMRRF DVDIILDLLV KLISFRIRFY DRFQIFSLQV LRKLEGQLGS EIIKNKIINS 720
721 PYGQAGITSV IDYMLECLSK NRNEACLAYE RPLPDLPSTI KPLAD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
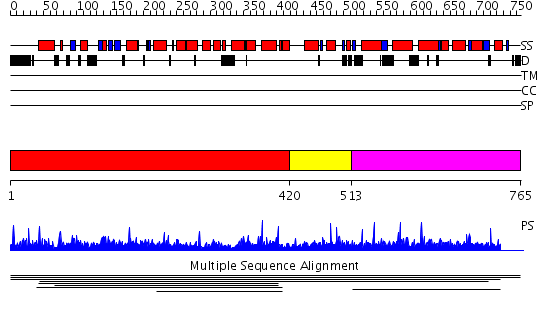
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..419] | 3.007894 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [420..512] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [513..765] | 1.004904 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)