













| General Information: |
|
| Name(s) found: |
BCH1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 724 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSQTSIPEV KEDVIGYALH QRRARVGQFQ DLGPPDLITL IKSLPSSSST TTATASANDN 60
61 GATSNINGQD PTTIVTELHS HDKLKGQIGT FFYCMGIDTS DPTSITIFAK KITDLFLDTP 120
121 QIWFGKKKHF HVSKISISSW NAFRKYDVNI IVHIPGTVQT YIINSDGEQS QLPSVAEASS 180
181 GRNSQDLNVN MIWAETFMSG IVRDIMIMKD NRADGESQNL VETLIFNPFT SGELEDVANN 240
241 FIKLFPLVYE KGVYLDAPTH VLNPSLTNNY LVETLVEIVR LTKSLEACRK MLKKLIEIHP 300
301 EAVIILIRVY FACDLEIDAV DLINEQLNSP SSFLADDSKT SHIQLIFKSE LLSIQSEFLL 360
361 DVKRDYKLAK EVAMEAVNCA PNEFKTWYLL TRIYIKLNDM SNALLSLNAC PMSQVKEKYV 420
421 LRRIAPITSD ENLHLPLPLD ASIEEISSLN PMDVQLEQKS ADPNLVNLSA SSLKSTFQLA 480
481 YKLLTEIVQI TGWEQLLKYR SKIFVMEDEY QGSTSSIDEA EVRGNDISKM RSKRLCERWL 540
541 DNLFMLLYED LKTYTDWQSE QLYFDAQNSK YHKLTVEWEL FGLCAKRLGH LPEAAKAFQI 600
601 GLSQRFSPVC AKNLLQFYID EHKRIRRDSV SANSELTSSQ ILSSINDIDS SIIDLVVKIC 660
661 CWNHRWYIEF SIILIDALSV AVQDMGITKV HNEIASRFSD PVAQLIDDNI LNFLKNFTND 720
721 TFDN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
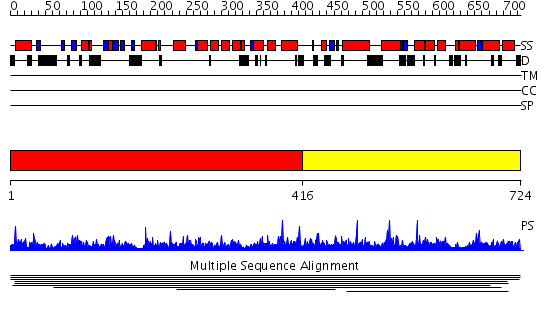
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..415] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [416..724] | 1.008926 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [637..724] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)