













| General Information: |
|
| Name(s) found: |
AVO1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1176 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTVTVLNEL RAQFLRVCPE KDQMKRIIKP YIPVDEFNTE QCLDSSIREL YMNSDGVSLL 60
61 PELESPPVSK DFMENYASLG KMRIMRENEG QKGKANQNLI GAEKTERDEE ETRNLQDKSA 120
121 KNTLIVEENG TLRYNPLNSS ASNSLLNDDD HTSGKHHKTS SKEDSYLNSS MEMQKKSSKR 180
181 SSLPFVRIFK SRRDHSNTSG NKNVMNTTNT RAKSSTLHPP GARHNKKGSK FDMNFDFDEN 240
241 LEEEDDDDDD DEEGDDIHSQ FFQLDDDFDA KGSGASPAHK GINGMSNNKN NTYTNNRNSI 300
301 SILDDRESSN GNIGSASRLK SHFPTSQKGK IFLTDNKNDG QKSDSLNANK GIHGDGSSAS 360
361 GNGSVSRDGL TETESNNISD MESYINEKDL DDLNFDTVTS NINKTVSDLG GHESTNDGTA 420
421 VMNRDSKDSR SNSNEFNAQN RDRITPGSSY GKSLLGSEYS EERYSNNDSS TMESGEMSLD 480
481 SDMQTNTIPS HSIPMSMQKY GIYHGDDDST LNNVFDKAVL TMNSSRHPKE RRDTVISGKE 540
541 PTSLTSSNRK FSVSSNLTST RSPLLRGHGR TSSTASSEHM KAPKVSDSVL HRARKSTLTL 600
601 KQDHSQPSVP SSVHKSSKEG NILIEKTTDY LVSKPKASQL SNMFNKKKKR TNTNSVDVLE 660
661 YFSFVCGDKV PNYESMGLEI YIQASKKYKR NSFTTKVRKS STIFEVIGFA LFLYSTEKKP 720
721 DNFEEDGLTV EDISNPNNFS LKIVDEDGEP FEDNFGKLDR KSTIQSISDS EVVLCKVDDA 780
781 EKSQNEIETP LPFETGGGLM DASTLDANSS HDTTDGTINQ LSFYKPIIGN EDDIDKTNGS 840
841 KIIDVTVYLY PNVNPKFNYT TISVLVTSHI NDILVKYCKM KNMDPNEYAL KVLGKNYILD 900
901 LNDTVLRLDG INKVELISKK DARELHLEKM KPDLKKPVLP TIQSNDLTPL TLEPLNSYLK 960
961 ADAGGAVAAI PENTKVTSKA KKISTKYKLG LAKQHSSSSV ASGSVSTAGG LANGNGFFKN1020
1021 KNSSKSSLHG TLQFHNINRS QSTMEHTPDT PNGVGDNFQD LFTGAYHKYK VWRRQQMSFI1080
1081 NKHERTLAID GDYIYIVPPE GRIHWHDNVK TKSLHISQVV LVKKSKRVPE HFKIFVRREG1140
1141 QDDIKRYYFE AVSGQECTEI VTRLQNLLSA YRMNHK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
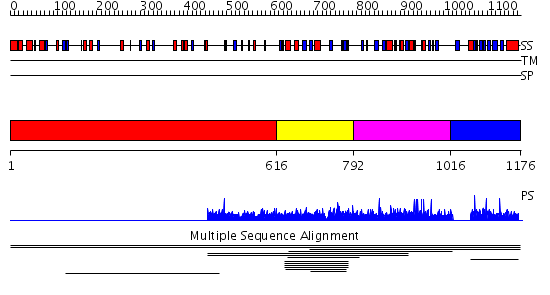
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..615] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [616..791] | 3.009964 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [792..1015] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1016..1176] | 1.004953 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [931..1051] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1052..1176] | 0.986 | Solution Structure of the Pleckstrin Homology Domain of Human Protein Kinase B beta (Pkb/Akt) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)