













| General Information: |
|
| Name(s) found: |
ARO80_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 950 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAKKRPSGN AAFELPKRRR TYQACISCRS RKVKCDLGPV DNPHDPPCAR CKRELKKCIF 60
61 SSNKGTSNDL PPNSINAISL PSLGKSKQEI QNDSTSPILS DVPLSRKGIS SEKSFKSEGM 120
121 KWKLELSSMQ NALEFLAQAA GTVAKEGAKE IIKEKSTTPK PLKSSLDATN KSATDEGLKR 180
181 LSKSDSTNTL YENTADMLNH TLNTNRKTSQ LMEEIGKVRP PPTRKIDDFD YIGPDSLLTK 240
241 EEAIELIEAF FLTMHPFFPN IPLQLHDPKE LAEYPILFCA ILTVSARYHP FDTLGLDNGE 300
301 DGMRHIEVHD KLWVYCQKLI SQTIWAEAST RSIGTVLAFI IFTEWNPRSI HYKWSDYAND 360
361 PELNNVNARG SKNISTRKDE EGLTGVGAIR RSDRMSWMLT GSAVRLAQDM GFIENSSKVF 420
421 IVTHISETTS AMNMNQRSLL AESFSVLNLN LGKIENDGNE SNEDYLGNEK FYLNEILPDE 480
481 ESKLRWKRVF ENSENDHDNE KNFLTDWERE FLNDEYVLYY SNKKDDTNLA QNHIPPFPLR 540
541 FSFAQRAKIE IIRILSIAYE TIYCEKNKRK LATTDQRHNL SVLSVFSPLI EGWLSNYREL 600
601 LVPLSDVPFS LADRKNKKQI FDNIDRINGE SIITDFNYCQ LYIFSLALQV DGKTSRLNMN 660
661 EIVTSARYVE LAYRSAKEIL SSAKRVSRQG MLKYMPVRWV IRIIRSIAFI VKCYLTLTGS 720
721 ELATNPDARN ILKLSAISVD ETFDIIRDTA VTLKEATPDE LHLCQRYAAI LMYLCTEMKL 780
781 RKKSYLERPP LLRDGTTPLE SNRESSLEGQ DLTKKPIFSK RIGYNKTETT FEPSERPLTE 840
841 EINSNSQNSN DTSSKGIVDP FVEQNNDITT ALLNNELFQG PSLSDEVTDW FGASEDIGLE 900
901 FVEPWTELIE QRYMQCGDGD NNNFENLYNL FVNSNNINND INNSRPITRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
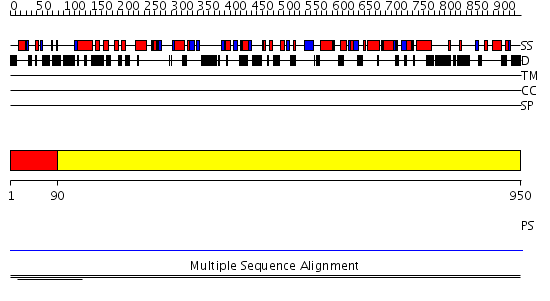
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 8.95 | Gal4; CD2-Gal4 | |
| 2 | View Details | [90..950] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [291..415] | 3.026992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [416..535] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [536..726] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [727..795] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [796..865] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [866..950] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)