













| General Information: |
|
| Name(s) found: |
ARGR2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 880 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGISSKNGPK KMGRAKTFTG CWTCRGRKVK CDLRHPHCQR CEKSNLPCGG YDIKLRWSKP 60
61 MQFDPYGVPI PQNSPATTTN LSGSVDEPQY QRRNIDFVRY DEEYVYHEDM DDELTMLHTP 120
121 PIEKISDNKT WIIKKFGVFK GTDKIDKQYA PRKKRNRKRV AKSLESSASI SLSSLPSSST 180
181 ISFPIRHIED KLRNKGHVKT GILSANDGVP PTPNLLDYDW NNLNITGYEW ISSELRDDAL 240
241 LSAVTLQGHH LGHTQPQEIS LEENSNVVSG EEHVNAKEHG CAFEADNQGS STLPNKAASA 300
301 NDKLYQQNLK LLFQKNSSNS EEPDPQALID DVFVNIEPRS LPASDLNKIT LAPPNEESRM 360
361 PKSMLELTSY SSDLPPELVD IIPKTDLTVH GLARFLLNHY FNNVADKMTV VVLEKNPWKT 420
421 LYFPRALMAL GDLAGLGQSS NSRNALLNAL LAVSCFHLQS KYPRNYKLQK YFLGLGIELR 480
481 NQASNFLRLC LNTKSSIPEK YKDVLTAILS MNSIDVVWGT MADCQDHLAL CEDFVESRMK 540
541 LRPNISEKTK TLHRIFSFLK LIQDSTALDK VRAKEIVILP SEEDDNYKPL DTSNATTSSS 600
601 EPRVDVVQEG LFREALNEND GKIHIEFVKE PITNVSADST PSSTTPPIFT NIATESYYNK 660
661 SDISKLVSKT DENIIGTDSL YGLPNSLILL FSDCVRIVRH NEYYNLTYLP VPRKFNELSL 720
721 NFEKRLLKWK SEWNFHQENS EGKSFINSTA EALYHHTMSF YFSLIIYYFT MARSLNCQFL 780
781 QNYVAKVLDH LNAMEELVDQ KKVKIVPLIW QGFMAGCACT DENRQQEFRR WAAKLAESGV 840
841 GSYWGARQVM LEVWRRRKED EPGDNWYSVY KDWEMNLMLS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
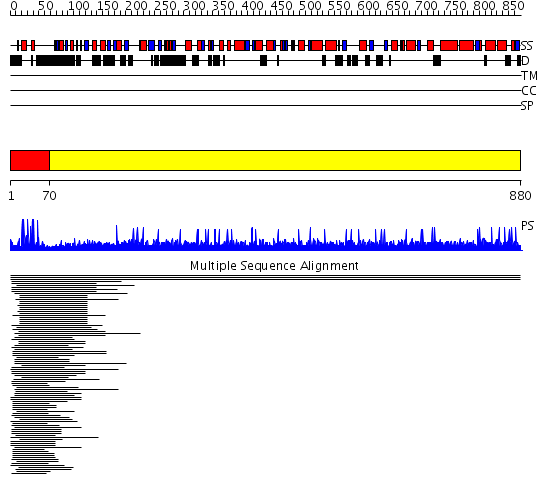
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | 46.68867 | PPR1 | |
| 2 | View Details | [70..880] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [205..295] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [296..355] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [356..580] | 1.021996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [581..671] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [672..880] | 2.016989 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)