













| General Information: |
|
| Name(s) found: |
ACON2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 789 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSSANRFYI KRHLATHANM FPSVSKNFQT KVPPYAKLLT NLDKIKQITN NAPLTLAEKI 60
61 LYSHLCDPEE SITSSDLSTI RGNKYLKLNP DRVAMQDASA QMALLQFMTT GLNQTSVPAS 120
121 IHCDHLIVGK DGETKDLPSS IATNQEVFDF LESCAKRYGI QFWGPGSGII HQIVLENFSA 180
181 PGLMMLGTDS HTPNAGGLGA IAIGVGGADA VDALTGTPWE LKAPKILGVK LTGKLNGWST 240
241 PKDVITKLAG LLTVRGGTGY IVEYFGEGVS TLSCTGMATI CNMGAEIGAT TSTFPYQEAH 300
301 KRYLQATNRA EVAEAADVAL NKFNFLRADK DAQYDKVIEI DLSAIEPHVN GPFTPDLSTP 360
361 ISQYAEKSLK ENWPQKVSAG LIGSCTNSSY QDMSRVVDLV KQASKAGLKP RIPFFVTPGS 420
421 EQIRATLERD GIIDIFQENG AKVLANACGP CIGQWNREDV SKTSKETNTI FTSFNRNFRA 480
481 RNDGNRNTMN FLTSPEIVTA MSYSGDAQFN PLTDSIKLPN GKDFKFQPPK GDELPKRGFE 540
541 HGRDKFYPEM DPKPDSNVEI KVDPNSDRLQ LLEPFKPWNG KELKTNVLLK VEGKCTTDHI 600
601 SAAGVWLKYK GHLENISYNT LIGAQNKETG EVNKAYDLDG TEYDIPGLMM KWKSDGRPWT 660
661 VIAEHNYGEG SAREHAALSP RFLGGEILLV KSFARIHETN LKKQGVLPLT FANESDYDKI 720
721 SSGDVLETLN LVDMIAKDGN NGGEIDVKIT KPNGESFTIK AKHTMSKDQI DFFKAGSAIN 780
781 YIGNIRRNE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
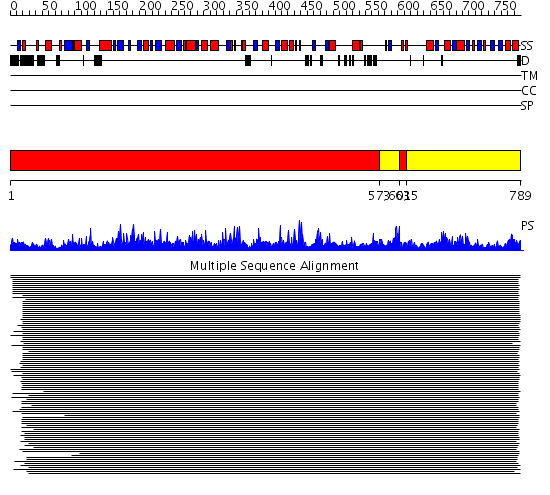
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..572] [603..614] |
11000.0 | Aconitase A, C-terminal domain; Aconitase A, N-terminal domain | |
| 2 | View Details | [573..602] [615..789] |
11000.0 | Aconitase A, C-terminal domain; Aconitase A, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)