













| General Information: |
|
| Name(s) found: |
TCPZB_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 535 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
cellular protein metabolic process
[IEA]
protein folding [IEA] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVRVLNPNA EVLNKSAALH MTINAAKGLQ DVLKSNLGPK GTIKMLVGGS GDIKLTKDGN 60
61 TLLKEMQIQN PTAIMIARTA VAQDDISGDG TTSTVIFIGE LMKQSERCID EGMHPRVLVD 120
121 GFEIAKRATL QFLDNFKTPV VMGDEVDKEI LKMVARTTLR TKLYEGLADQ LTDIVVNSVL 180
181 CIRKPEEAID LFMVEIMHMR HKFDVDTRLV EGLVLDHGSR HPDMKRRAEN CHILTCNVSL 240
241 EYEKSEINAG FFYSNAEQRE AMVTAERRSV DERVKKIIEL KKKVCGDNDN FVVINQKGID 300
301 PPSLDLLARE GIIGLRRAKR RNMERLVLAC GGEAVNSVDD LTPESLGWAG LVYEHVLGEE 360
361 KYTFVEQVKN PNSCTILIKG PNDHTIAQIK DAVRDGLRSV KNTIEDECVV LGAGAFEVAA 420
421 RQHLLNEVKK TVQGRAQLGV EAFANALLVV PKTLAENAGL DTQDVIISLT SEHDKGNVVG 480
481 LNLQDGEPID PQLAGIFDNY SVKRQLINSG PVIASQLLLV DEVIRAGRNM RKPTA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
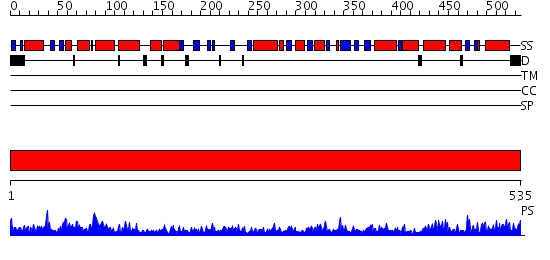
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..535] | 143.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)