













| General Information: |
|
| Name(s) found: |
PNP_LISMO
[Swiss-Prot]
|
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Listeria monocytogenes EGD-e |
| Length: | 723 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEKQVFSTE WAGKTLSVEV GQLAKQASGA ALIRYGDTVV LTAAVGSKKP RPGDFFPLTV 60
61 NYEEKMYSVG KVPGGFLKRE GRPSDRATLT ARLIDRPIRP LFAEGFRNEV QITSTVFSVD 120
121 QDCSPEMAAM LGSSVALVIS DIPFEGPIAG VDVGRIDGKY VINPTIEQAE KSDISLTVAG 180
181 TYDAINMVEA GAKEVSEEAM LEAIMFGHEE IKRLCEFQQQ IIAAVGKEKR EIELFVSDPE 240
241 LEAEVKAASE GKMKTAIKTE EKKAREAAIE EVKEEILESY KAKELENESE ILSEVAHILE 300
301 MIEKDEMRRL ISQDKIRPDG RKVNEIRPLS SEVGMLPRVH GSGLFTRGQT QALSVCTLAP 360
361 LREHQIIDGL GTEEYKRFMH HYNFPQFSVG ETGPRRAPGR REIGHGALGE RALQYVIPSE 420
421 EEFPYTIRLV SEVLESNGSS SQASICGSTL AMLDAGVPIK APVAGIAMGL VKLGDDYTIL 480
481 SDIQGMEDHF GDMDFKVAGT KDGITALQMD IKIDGLSRQI LDEALTQAKE GRLHILEHLT 540
541 STISAPREEL SAYAPKIITL NIKPEKIKDV IGPGGKQINA IIDETGVKID IEQDGTVYIA 600
601 SQDQAMNRKA IAIIEDIVRE VEVGEVYTGK VRRIEKFGAF VELFKGTDGL VHISELAHER 660
661 VGKVEDILKL GDEVTVKVIE VDQQGRVNLS RKALLEKKEQ PEGDKKPQAE KKFYPKTKKP 720
721 ESK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
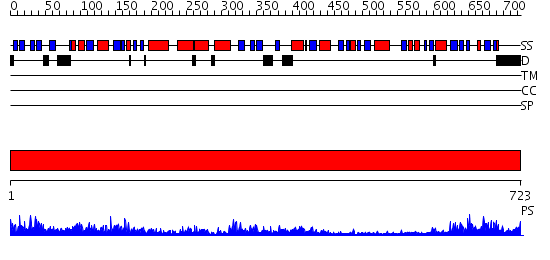
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..723] | 140.0 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)