













| General Information: |
|
| Name(s) found: |
TGH_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 930 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
[NOT]auxin polar transport
[IMP]
phloem or xylem histogenesis [IMP] RNA processing [ISS][TAS] [NOT]auxin mediated signaling pathway [IMP] |
| Molecular Function: |
RNA binding
[ISS][TAS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSDEEDFVF HGTPIEREEE IASRKKKAVA GASGNLRTLP AWKQEVTDEE GRRRFHGAFT 60
61 GGYSAGYYNT VGSKEGWAPQ SFTSSRQNRA GARKQSISDF LDEDEKADME GKSLSASSQF 120
121 DTFGFTAAEH SRKHAEKEQH ERPSAIPGPV PDELVAPVSE SIGVKLLLKM GWRRGHSIKE 180
181 VRASSDARRE ARKAFLAFYT DENTKETPDS LVSETEVETS LGEDIKISES TPVYVLNPKQ 240
241 DLHGLGYDPF KHAPEFREKK RSRMSANKEV GFRKPLSMKE SLFGPKSGKI APGFGIGALE 300
301 ELDVEDEDVY AGYDFDQTYV IEDEQPARQS NDNRLRLTSK EHDVLPGFGA AKNSDYSMER 360
361 FNPPIIPKDF VARHKFSGPL EAETKPTVSA PPEVPPPADN NLKLLIEGFA TFVSRCGKLY 420
421 EDLSREKNQS NQLFDFLREG NGHDYYARRL WEEQQKRKDQ SKLTLDVKVS PTVQKMTAET 480
481 RGSLLGEKPL QRSLKETDTS ASSGGSFQFP TNLSDTFTKS ASSQEAADAV KPFKDDPAKQ 540
541 ERFEQFLKEK YKGGLRTTDS NRVNSMSESA RAQERLDFEA AAEAIEKGKA YKEVRRATEQ 600
601 PLDFLAGGLQ FTSGGTEQIK DTGVVDMKSS KTYPKREEFQ WRPSPLLCKR FDLPDPFMGK 660
661 LPPAPRARNK MDSLVFLPDT VKAASARQVS ESQVPKKETS IEEPEVEVEV ENVERPVDLY 720
721 KAIFSDDSED DEDQPMNGKI QEGQEKKNEA AATTLNRLIA GDFLESLGKE LGFEVPMEEE 780
781 IKSRSKPEDS SDKRLDRPGL KEKVEEKTSS LTLGSEEEKS RKKREKSPGK RSGGNDLSSS 840
841 ESSGDERRRK RYNKKDRHRN DSESDSSSDY HSRDKQGSRS RSKRRESSRE KRSSHKKHSK 900
901 HRRTKKSSSS RYSSDEEQKE SRREKKRRRD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
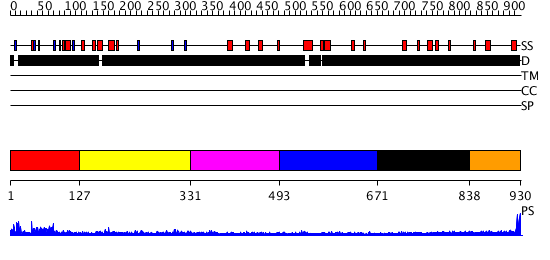
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 52.657577 | No description for PF07713.5 was found. No confident structure predictions are available. | |
| 2 | View Details | [127..330] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [331..492] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [493..670] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [671..837] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [838..930] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)