













| General Information: |
|
| Name(s) found: |
OBGC_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 681 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
embryonic development
[IMP]
embryonic development ending in seed dormancy [NAS] response to light stimulus [IEP] |
| Molecular Function: |
GTPase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASISINCFF TPQALARPSR TRKIFAKPDK VSGRTRSPRK TKLQREVELK SRGGDKLQPV 60
61 SDAGGEATTY TRLPPREDYY DVSLLSSSYL KVSEEVKLSE SNVARVEEKI ETLREIEEEE 120
121 KEKEVKSYDD DDIWGNYRRL DVFEGSSRSI DEDDEDWEDE VFEYGDETDA EKDDSEGSEL 180
181 KDGEVLCFSG EEEEDEIGVK EKGVPAVMRC FDRAKIYVRA GDGGNGVVAF RREKFVPFGG 240
241 PSGGDGGRGG NVYVEVDGSM NSLLPFRKSV HFRAGRGEHG RGKMQSGAKG DNVVVKVAPG 300
301 TVVRQAREVG SEVEGEEGEE KEVLLELLHP GQRALLLPGG RGGRGNASFK SGMNKVPRIA 360
361 ENGEEGPEMW LDLELKLVAD VGIVGAPNAG KSTLLSVISA AQPTIANYPF TTLLPNLGVV 420
421 SFDYDSTMVV ADLPGLLEGA HRGFGLGHEF LRHTERCSAL VHVVDGSAPQ PELEFEAVRL 480
481 ELELFSPEIA EKPYVVAYNK MDLPDAYEKW PMFQETLRAR GIEPFCMSAV QREGTHEVIS 540
541 SVYELLKKYR AANAEPKALF DQANENLDHV AKKIDKERRA AINEFEVFRD SGTRAWHVVG 600
601 AGLQRFVQMT NWRYMDSDKR FQHVLDACGV NKSLKNMGVK EGDTVIVGEM ELIWHDSANG 660
661 SSRPTDSNKT STDSVRWPQW K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
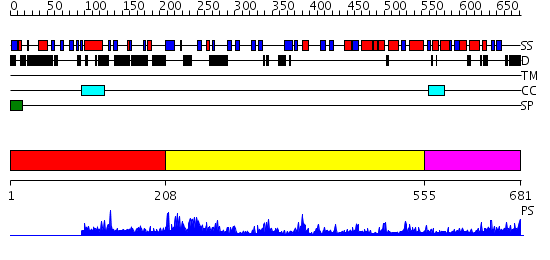
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [208..554] | 81.0 | Obg GTP-binding protein N-terminal domain; Obg GTP-binding protein C-terminal domain | |
| 3 | View Details | [555..681] | 50.69897 | IF2, IF1, and tRNA fitted to cryo-EM data OF E. COLI 70S initiation complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.53 |
Source: Reynolds et al. (2008)