













| General Information: |
|
| Name(s) found: |
PID2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 525 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid phosphorylation
[ISS]
cotyledon development [IGI] |
| Molecular Function: |
ATP binding
[ISS]
protein kinase activity [ISS] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANSSIFYKD NESDYESSTV GPDSSRRTSW LSSSFTASPS CSSISHLSNH GLNSYNQSKP 60
61 HKANQVAWEA MARLRRCCGR AVGLEHFRLL KRLGSGDIGS VYLCQIRGSP ETAFYAMKVV 120
121 DKEAVAVKKK LGRAEMEKKI LGMLDHPFCP TLYAAFEASH YSFLVMEYCP GGDLYAVRLR 180
181 QPSKRFTISS TRFYAAETLV ALEYLHMMGI VYRDLKPENV LIREDGHVML SDFDLSFKCD 240
241 VVPQFLSDND RDRGHQEDDD DISIRRKCST PSCTTTPLNP VISCFSPTSS RRRKKNVVTT 300
301 TIHENAAGTS DSVKSNDVSR TFSRSPSSCS RVSNGLRDIS GGCPSIFAEP INARSKSFVG 360
361 THEYLAPEVI SGQGHGSAVD WWTYGIFLYE MIFGRTPFKG DNNEKTLVNI LKAPLTFPKV 420
421 IVNSPKEYED MVNAQDLIIK LLVKNPKKRL GSLKGSIEIK RHEFFEGVNW ALIRSIKPPW 480
481 VPKEETSHKT KGDNRSVNYY LPPRFMMSRK ERNEPYHVSN YFDYF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
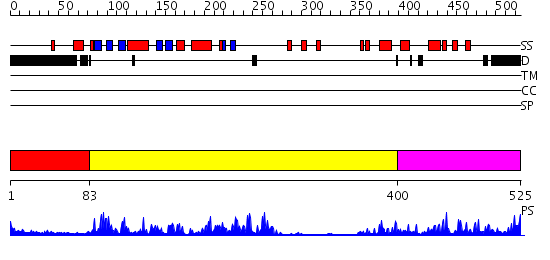
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 53.522879 | CHICKEN SRC TYROSINE KINASE | |
| 2 | View Details | [83..399] | 67.30103 | crystal structure of an inactive Akt2 kinase domain | |
| 3 | View Details | [400..525] | 4.2 | No description for 2jdoA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)