













| General Information: |
|
| Name(s) found: |
nth /
EG10662
[EcoGene]
|
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli str. K-12 substr. MG1655 |
| Length: | 211 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
DNA repair
[IEA]
base-excision repair [IEA] metabolic process [IEA] response to DNA damage stimulus [IEA] |
| Molecular Function: |
DNA binding
[IEA]
protein binding [IPI metal ion binding [IEA] iron ion binding [IEA] catalytic activity [IEA] endonuclease activity [IEA] lyase activity [IEA] DNA-(apurinic or apyrimidinic site) lyase activity [IEA] 4 iron, 4 sulfur cluster binding [IEA] iron-sulfur cluster binding [IEA] hydrolase activity, acting on glycosyl bonds [IEA] hydrolase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKAKRLEIL TRLRENNPHP TTELNFSSPF ELLIAVLLSA QATDVSVNKA TAKLYPVANT 60
61 PAAMLELGVE GVKTYIKTIG LYNSKAENII KTCRILLEQH NGEVPEDRAA LEALPGVGRK 120
121 TANVVLNTAF GWPTIAVDTH IFRVCNRTQF APGKNVEQVE EKLLKVVPAE FKVDCHHWLI 180
181 LHGRYTCIAR KPRCGSCIIE DLCEYKEKVD I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
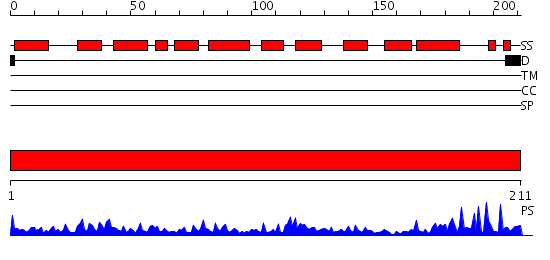
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..211] | 70.0 | REFINEMENT OF THE NATIVE STRUCTURE OF ENDONUCLEASE III TO A RESOLUTION OF 1.85 ANGSTROM |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)