













| General Information: |
|
| Name(s) found: |
gi|19114900
[NCBI NR]
gi|13624754 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 570 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSVEKASKA CELCRRKKIR CNRELPSCQN CIVYQEECHY SKRLKRSYSA TKKKNGNPVL 60
61 ESAIPSLSPS PSIENGSAML NSDITSLSNR IFKVEEKLDL ILSLLKNSSE PLDRTERKDF 120
121 PSLAMQIRDA NSLVNTKLKE YSRRFELPSQ KTSFDDLFSS TFPNFDAAFK DIPDKEWAFE 180
181 NVQWYFRYIN CWWPVFYEKD FMDEYECLYR DRNQVKGAWL VSFYSVLALA ASRSKAGKDQ 240
241 KLAESFFSTS WYLIQKPGFF LTPQLEKIQA LLIMIQFAAH VSLHTLCKAL CGQACLMIRD 300
301 LNLHRESANA DFSNKDAELR RRVFWICYIF EITTSLVFGT PSVLSDMDID CEHPNYEYGR 360
361 YFSEMPTGDL IFSSEVSLTI LKNEVRTKVY SRTNTSNARN REKAIWQIHE KLLCWERALP 420
421 IELRQYFIAL TENAQIYEEL DFEKQRLFSA CIEVYLSYCN TLIFLHRLNE SVEGANICLD 480
481 TARRAINVLK FFFIIPIAKN VCYLWVFLYC PFTPFLVLFS NIVNGKEPST DIAFEDLNRM 540
541 YSVNRFFVKL RDIGGDLAEK LASVTENFIH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
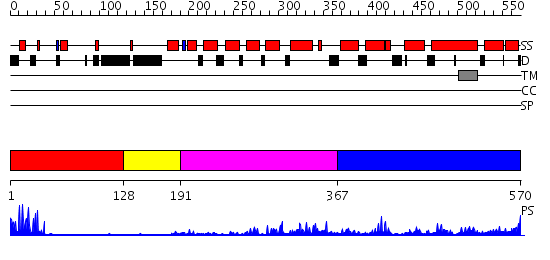
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 4.69897 | Gal4; CD2-Gal4 | |
| 2 | View Details | [128..190] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [191..366] | 32.619789 | No description for PF04082.9 was found. Confident ab initio structure predictions are available. | |
| 4 | View Details | [367..570] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.58 |
Source: Reynolds et al. (2008)