













| General Information: |
|
| Name(s) found: |
PLSP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 291 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
membrane [ISS] plastid envelope [IDA] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
proteolysis
[ISS]
thylakoid membrane organization [IMP] protein maturation [IMP] |
| Molecular Function: |
peptidase activity
[IMP][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMVMISLHFS TPPLAFLKSD SNSRFLKNPN PNFIQFTPKS QLLFPQRLNF NTGTNLNRRT 60
61 LSCYGIKDSS ETTKSAPSLD SGDGGGGDGG DDDKGEVEEK NRLFPEWLDF TSDDAQTVFV 120
121 AIAVSLAFRY FIAEPRYIPS LSMYPTFDVG DRLVAEKVSY YFRKPCANDI VIFKSPPVLQ 180
181 EVGYTDADVF IKRIVAKEGD LVEVHNGKLM VNGVARNEKF ILEPPGYEMT PIRVPENSVF 240
241 VMGDNRNNSY DSHVWGPLPL KNIIGRSVFR YWPPNRVSGT VLEGGCAVDK Q |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
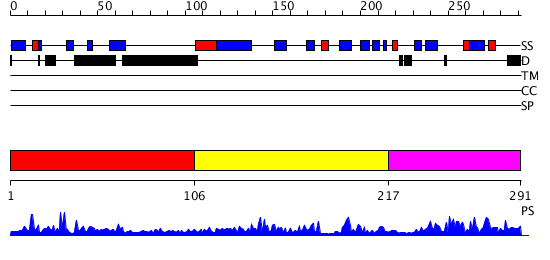
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [106..216] | 19.221849 | Crystal structure of Escherichia coli type I signal peptidase in complex with a lipopeptide inhibitor | |
| 3 | View Details | [217..291] | 4.23 | Crystal structure of Escherichia coli type I signal peptidase in complex with a lipopeptide inhibitor |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)