













| General Information: |
|
| Name(s) found: |
ODP22_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 539 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrion [IDA] chloroplast envelope [IDA] |
| Biological Process: |
metabolic process
[IEA]
pyruvate metabolic process [IEA] |
| Molecular Function: |
copper ion binding
[IDA]
dihydrolipoyllysine-residue acetyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASRIINHSK KLKHVSALLR RDHAVAVRCF SNSTHPSLVG REDIFKARLN YSSVERISKC 60
61 GTGNVTMLSG ISTTSTKLSS PMAGPKLFKE FISSQMRSVR GFSSSSDLPP HQEIGMPSLS 120
121 PTMTEGNIAR WLKKEGDKVA PGEVLCEVET DKATVEMECM EEGFLAKIVK EEGAKEIQVG 180
181 EVIAITVEDE DDIQKFKDYT PSSDTGPAAP EAKPAPSLPK EEKVEKPASA PEAKISKPSS 240
241 APSEDRIFAS PLARKLAEDN NVPLSSIKGT GPEGRIVKAD VEDFLASGSK ETTAKPSKQV 300
301 DSKVPALDYV DIPHTQIRKV TASRLAFSKQ TIPHYYLTVD TCVDKMMGLR SQLNSFQEAS 360
361 GGKRISVNDL VIKAAALALR KVPQCNSSWT DEYIRQFKNV NINVAVQTEN GLYVPVVKDA 420
421 DKKGLSTIGE EVRFLAQKAK ENSLKPEDYE GGTFTVSNLG GPFGIKQFCA VINPPQAAIL 480
481 AIGSAEKRVV PGTGPDQYNV ASYMSVTLSC DHRVIDGAIG AEWLKAFKGY IETPESMLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
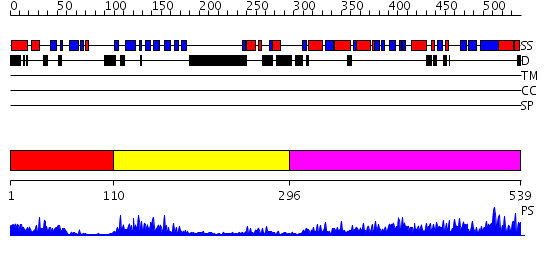
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 12.39794 | No description for 2qf7A was found. | |
| 2 | View Details | [110..295] | 25.154902 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 3 | View Details | [296..539] | 85.69897 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)