













| General Information: |
|
| Name(s) found: |
GCS2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 789 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
oligosaccharide metabolic process
[IEA]
|
| Molecular Function: |
alpha-glucosidase activity
[ISS]
catalytic activity [IEA] mannosyl-oligosaccharide glucosidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTGLSPSGDI KPLLDDESQR PRVITPFPSP KLTDLSMFQG EHKESLYWGT YRPQMYFGVR 60
61 ARTPKSLVAG LMWLVMKDGK PVMRHFCENS NDLKSFGWKE HNGRDFGRQE LFEQNMVLET 120
121 SFVKSEEGSL GYGGDWSIRI NVQNMLNDDE VKRSTHLFFY LADEGCNGVN LGKDVLRLKE 180
181 SSVLASGSRQ DVGNWQMHLK SQNHLETHYC GFKTPDIVNL SGLVQQNLTA QEEKSGLLQL 240
241 SDSSEDSSNI YVFQISTTNQ STIDIAFVSG IKEETSNMEN RIMSLTGLPL SSLLEEKHIA 300
301 FNAKFNECFN LSDKLDPETL VVGKAAIGNM LGGIGYFYGQ SKIHFPKSPL AKSEDDFLLY 360
361 WPAELYTAVP SRPRFPRGFL WDEGFHQLLI WRWDVHITLE IVGNWLDLMN IDGWIPREQV 420
421 LGAEALSKIP KQYVVQFPSN GNPPTLLLVI RDLINGIRTE KFNEADRDKI LSFLDRAFVR 480
481 LDAWFKWFNT SQIGKEKGSY YWHGRDNITN RELNPQSLSS GLDDYPRASH PNEDERHVDL 540
541 RCWMYLAADS MNSIQEFMGK NDILVTEDYS SIAKLLSDFT LLNQMHYDKD HGAYLDFGNH 600
601 TEEVRLVWKE VIHEDGHLSR ELVRETYGKP ELRLVPHIGY VSFFPFMFRI IPADSSILDK 660
661 QLDLISNGNI VWSDYGLLSL AKTSSLYMKY NSEHQAPYWR GAIWMNMNYM ILSSLHHYST 720
721 VDGPYNSKAR TIYEELRSNL IRNVVRNYDQ TGIIWEHYDQ TKGTGEGARV FTGWSALILL 780
781 IMSEEYPLF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
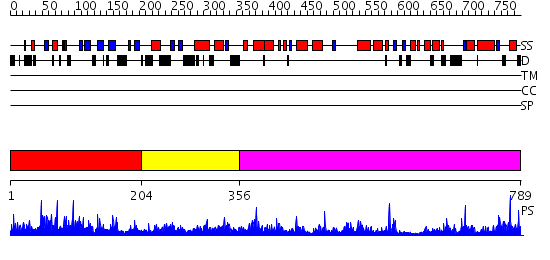
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..203] | 0.969 | Afx (Foxo4) | |
| 2 | View Details | [204..355] | 1.126972 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [356..789] | 63.39794 | No description for 2jf4A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)