













| General Information: |
|
| Name(s) found: |
SLK2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 816 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
embryonic development
[IMP]
gynoecium development [IMP] ovule development [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSTSGIFF QGDDESQSFI NSHLTSSYGN SSNSAPGCGG PTGGYHNLSM VSGDMHNPVM 60
61 MSVSTPGPSA GASSLVTDAN SGLSGGGPHL QRSASINNES YMRLPASPMS FSSNNISISG 120
121 SSVVDGSTVV QRHDPSVQLG GSSATSLPTS QTNQIPLSMA RRASESFFQD PNNLTQARKK 180
181 PRLDSKQDDA LQQQILRQWL QRQDILQQQQ QQQQQGQNPQ FQILLQQQKL RQQQQYLQSL 240
241 PPLQRVQLQQ QQQVQQQQQL QQQHQQQQQQ LQQQGMQMQL TGGPRPYENS VCARRLMQYL 300
301 YHQRQRPSES SIVYWRKFVT EYFSPRAKKR WCLSHYDNVG HSALGVSPQA ATDEWQCDLC 360
361 GSKSGRGFEA TFDVLPRLNE IKFASGVLDE LLYLGVPSER RYGSGIMVLE YGKAVQESVY 420
421 EHIRVVREGH LRIIFSQELK ILSWEFCTRR HEELLPRRLV APQVNQLLQV AEKCQSTIDQ 480
481 SGSDGIHQQD LQANSNMVMA AGRQLAKSLE SHSLNDLGFS KRYVRCLQIS EVVSSMKDMI 540
541 DFCRDQKVGP IEALKSYPYR MKAGKPQMQE MEQLAAARGL PPDRNSLNKL MALRNSGINI 600
601 PMNNMSGQGS LPGSAQAAAF ALTNYQSMLM KQNHLNSDLN NTTIQQEPSR NRSASPSYQG 660
661 TSPLLPGFVH SPSISGVSSH LSPQRQMPSS SYNGSTQQYH QQPPSCSSGN QTLEQQMIHQ 720
721 IWQQMANSNG GSGQQQQSLS GQNMMNCNTN MGRNRTDYVP AAAETPSTSN RFRGIKGLDQ 780
781 SQNLEGIISN TSLNFGNNGV FSNEVDESMG GYSWKS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
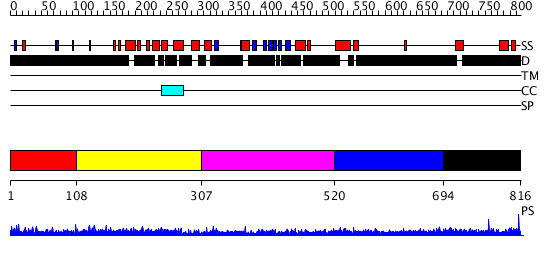
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [108..306] | 4.69897 | Sec24 | |
| 3 | View Details | [307..519] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [520..693] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [694..816] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)