













| General Information: |
|
| Name(s) found: |
SBT4C_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 736 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] apoplast [IDA] |
| Biological Process: |
negative regulation of catalytic activity
[IEA]
proteolysis [IEA][ISS] |
| Molecular Function: |
identical protein binding
[IEA]
serine-type endopeptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANLAASTCL YSWLLVLLLS SVSAIIDEDT QVYIVYMGSL SSRADYIPTS DHMSILQQVT 60
61 GESSIEGRLV RSYKRSFNGF AARLTESERT LIAEIEGVVS VFPNKILQLH TTTSWDFMGV 120
121 KEGKNTKRNL AIESDTIIGV IDTGIWPESK SFSDKGFGPP PKKWKGVCSG GKNFTCNNKL 180
181 IGARDYTSEG TRDTSGHGTH TASTAAGNAV KDTSFFGIGN GTVRGGVPAS RIAAYKVCTD 240
241 SGCSSEALLS SFDDAIADGV DLITISIGFQ FPSIFEDDPI AIGAFHAMAK GILTVSSAGN 300
301 SGPKPTTVSH VAPWIFTVAA STTNRGFITK VVLGNGKTLA GRSVNAFDMK GKKYPLVYGK 360
361 SAASSACDAK TAALCAPACL NKSRVKGKIL VCGGPSGYKI AKSVGAIAII DKSPRPDVAF 420
421 THHLPASGLK AKDFKSLVSY IESQDSPQAA VLKTETIFNR TSPVIASFSS RGPNTIAVDI 480
481 LKPDITAPGV EILAAFSPNG EPSEDDTRRV KYSVFSGTSM ACPHVAGVAA YVKTFYPRWS 540
541 PSMIQSAIMT TAWPVKAKGR GIASTEFAYG AGHVDPMAAL NPGLVYELDK ADHIAFLCGM 600
601 NYTSKTLKII SGDTVKCSKK NKILPRNLNY PSMSAKLSGT DSTFSVTFNR TLTNVGTPNS 660
661 TYKSKVVAGH GSKLSIKVTP SVLYFKTVNE KQSFSVTVTG SDVDSEVPSS ANLIWSDGTH 720
721 NVRSPIVVYI MVVDEA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
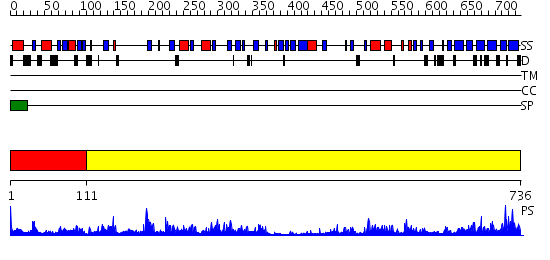
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] | 44.30103 | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin | |
| 2 | View Details | [111..736] | 50.69897 | Structure of C5a peptidase- a key virulence factor from Streptococcus |
| Source: Reynolds et al. 2008. Manuscript submitted |

|