













| General Information: |
|
| Name(s) found: |
ISOA3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 764 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] chloroplast starch grain [IDA] |
| Biological Process: |
carbohydrate metabolic process
[ISS]
starch catabolic process [IMP] |
| Molecular Function: |
alpha-amylase activity
[ISS]
isoamylase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLTSPSSSST YDPFSSNFSP SLTNAFSSSF TIPMGLKLSR RVTRARIFSR KIKDRSTLKV 60
61 TCRRAHERVV EEEASTMTET KLFKVSSGEV SPLGVSQVDK GINFALFSQN ATSVTLCLSL 120
121 SQSGKDDTDD DGMIELVLDP SVNKTGDTWH ICVEDLPLNN VLYGYRVDGP GEWQQGHRFD 180
181 RSILLLDPYA KLVKGHSSFG DSSQKFAQFY GTYDFESSPF DWGDDYKFPN IPEKDLVIYE 240
241 MNVRAFTADE SSGMDPAIGG SYLGFIEKIP HLQDLGINAV ELLPVFEFDE LELQRRSNPR 300
301 DHMVNTWGYS TVNFFAPMSR YASGEGDPIK ASKEFKEMVK ALHSAGIEVI LDVVYNHTNE 360
361 ADDKYPYTTS FRGIDNKVYY MLDPNNQLLN FSGCGNTLNC NHPVVMELIL DSLRHWVTEY 420
421 HVDGFRFDLA SVLCRATDGS PLSAPPLIRA IAKDSVLSRC KIIAEPWDCG GLYLVGKFPN 480
481 WDRWAEWNGM YRDDVRRFIK GDSGMKGSFA TRVSGSSDLY QVNQRKPYHG VNFVIAHDGF 540
541 TLRDLVSYNF KHNEANGEGG NDGCNDNHSW NCGFEGETGD AHIKSLRTRQ MKNFHLALMI 600
601 SQGTPMMLMG DEYGHTRYGN NNSYGHDTSL NNFQWKELDA KKQNHFRFFS EVIKFRHSHH 660
661 VLKHENFLTQ GEITWHEDNW DNSESKFLAF TLHDGIGGRD IYVAFNAHDY FVKALIPQPP 720
721 PGKQWFRVAD TNLESPDDFV REGVAGVADT YNVAPFSSIL LQSK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
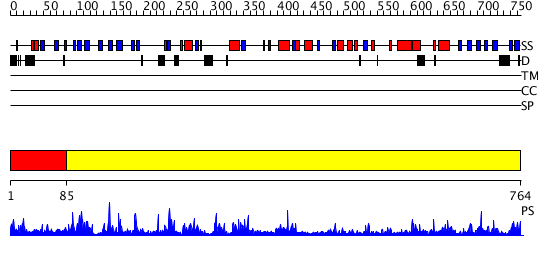
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [85..764] | 89.154902 | STRUCTURE OF PSEUDOMONAS ISOAMYLASE |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)