













| General Information: |
|
| Name(s) found: |
FAS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 815 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin assembly complex
[IPI]
|
| Biological Process: |
cell proliferation
[IMP]
heterochromatin formation [IMP] leaf development [IMP] DNA recombination [IEP][IMP] double-strand break repair via homologous recombination [IMP] pollen development [IGI] positive regulation of cell cycle [IGI] regulation of meristem structural organization [IMP] chromatin organization [IDA] nucleosome assembly [IDA] trichome differentiation [IMP] |
| Molecular Function: |
histone binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEVSTVNEN ENRKTMIEPK KLNKRKREPT AIENLTSEEK ESQISSLNLE MKGLFDYFRE 60
61 VMDKSKRTDL FSGFSECSSL NSMVALLMEE MSLPLSKLVD EIYLKLKEKT ESVTMVAVKS 120
121 AVVSVGQRVS YGVLNVDADV LEDDSESCLW CWETRDLKIM PSSVRGVLKL RRTCRKKIHE 180
181 RITAVSAMLA ALQREETEKL WRSDLSKAAE KLGKILSEVD IRSFMDNMMQ KNSSEMAEKD 240
241 SKREEKLLLK QLEKNRCEAE KEKKRMERQV LKEKLQQEKE QKLLQKAIVD ENNKEKEETE 300
301 SRKRIKKQQD ESEKEQKRRE KEQAELKKQL QVQKQASIME RFLKKSKDSS LTQPKLPSSE 360
361 VTAQELSCTK HENEIGKVVQ AIDNAFSTTC EATVDDIRRE HFASWRQLGH LLSSSKKHWG 420
421 MRRQPKSELF PKLKLSTNSG VTSDGEPNME KQGDGCEENN FDGRQCKPSS SNRKKSRRVK 480
481 QLLQFDKSCR PGFYGIWPSQ SQVVKPRRPL QKDPELDYEV DSDEEWEEEE AGESLSDCEK 540
541 DEDESLEEGC SKADDEDDSE DDFMVPDGYL SEDEGVQVDR MDIDPSEQDA NTTSSKQDQE 600
601 SPEFCALLQQ QKHLQNLTDH ALKKTQPLII CNLTHEKVSL LAAKDLEGTQ KVEQICLRAL 660
661 MVRQFPWSSL IEISINDIQD EDQEASKFSC SQSTPPSNSK AKIIPDSDLL TVVSTIQSCS 720
721 QGINRVVETL QQKFPDVPKT KLRQKVREIS DFEDSRWQVK KEVLTKLGLS PSPDKGGKRL 780
781 PKTISTFFSK RCLPPSTKPQ PAVEDAAERL ENENA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
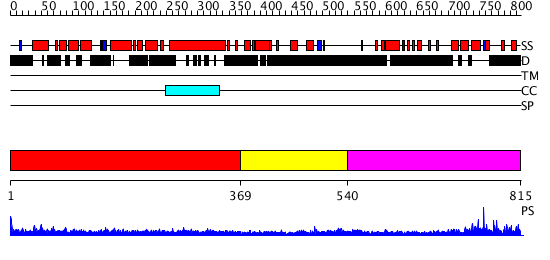
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..368] | 23.522879 | Heavy meromyosin subfragment | |
| 2 | View Details | [369..539] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [540..815] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)