













| General Information: |
|
| Name(s) found: |
gi|254852103
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Listeria monocytogenes FSL R2-503 |
| Length: | 608 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKEEMNARQ KKIRNFSIIA HIDHGKSTLA DRILEQTGAL THREMKNQLL DSMDLERERG 60
61 ITIKLNAVQL KYKAKDGETY IFHLIDTPGH VDFTYEVSRS LAACEGAILV VDAAQGIEAQ 120
121 TLANVYLALD NDLEILPVIN KIDLPAADPE RVREEIEDVI GLDASDAVLA SAKSGIGIED 180
181 ILEQIVEKVP EPSGDVNKPL KALIFDSVFD AYRGVIANIR IMDGVVKAGD RIKMMSNGKE 240
241 FEVTEVGVFS PKATPRDELL VGDVGYLTAA IKNVGDTRVG DTITLANNPA EEALDGYRKL 300
301 NPMVYCGLYP IDSSKYNDLR DALEKLELND SALQFEAETS QALGFGFRCG FLGLLHMEII 360
361 QERIEREFNI DLITTAPSVI YHVNLTDGSN IVVDNPAEMP EPGVIESVEE PYVKATVMVP 420
421 NDYVGAVMEL AQNKRGNFIT MEYLDDIRVS IVYEIPLSEI VYDFFDQLKS STKGYASFDY 480
481 ELIGYKASKL VKMDILLNAE KVDALSFIVH RDFAYERGKI IVEKLKELIP RQQFEVPIQA 540
541 AIATKIVSRS TIKALRKNVL AKCYGGDVSR KRKLLEKQKE GKKRMKQIGS VEVPQEAFMA 600
601 ILKMDESK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
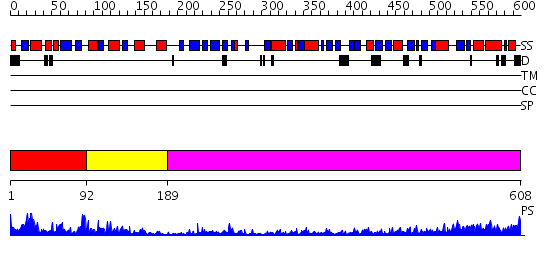
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | 59.522879 | Elongation factor eEF-1alpha, domain 2; Elongation factor eEF-1alpha, C-terminal domain; Elongation factor eEF-1alpha, N-terminal (G) domain | |
| 2 | View Details | [92..188] | 3.76 | Initiation factor IF2/eIF5b, domains 2 and 4; Initiation factor IF2/eIF5b, domain 3; Initiation factor IF2/eIF5b, N-terminal (G) domain | |
| 3 | View Details | [189..608] | 55.09691 | Ribosomal elongation factor G (EF-G) Fusidic acid resistant mutant T84A. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)