













| General Information: |
|
| Name(s) found: |
gi|30680771
[NCBI NR]
gi|21928125 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 471 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA repair
[IEA]
mRNA capping [IEA][ISS] DNA replication [IEA] protein amino acid dephosphorylation [IEA] DNA recombination [IEA] mRNA processing [IEA][ISS] dephosphorylation [IEA] |
| Molecular Function: |
ATP binding
[IEA]
DNA ligase (ATP) activity [IEA] protein tyrosine phosphatase activity [IEA] phosphatase activity [IEA] mRNA guanylyltransferase activity [IEA][ISS] polynucleotide 5'-phosphatase activity [IEA] protein tyrosine/serine/threonine phosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVATMDLNAS PQPEEDDEPY VRHLEDYSSR DERIESAVEI ARREREERKK RMRYDKPTHN 60
61 SQPVFRDQYY QNRNTKAYDR YKIPQGWLDC PPSGNEIGFL VPSKVPLNES YNNHVPPGSR 120
121 YSFKQVIHNQ RIAGRKLGLV IDLTNTTRYY STTDLKKEGI KHVKIACKGR DAVPDNVSVN 180
181 AFVNEVNQFV LNLKHSKKYI LVHCTHGHNR TGFMIVHYLM RSGPMNVTQA LKIFSDARPP 240
241 GIYKPDYIDA LYSFYHEIKP ESVICPSTPE WKRSTELDLN GEALPDDEDD DGGPAGPVQG 300
301 FQEESHQVDV KMSNDDVLGD EIPPDQEEGY RQFFYRMLSL NIGGRGCSQF PGSHPVSLNR 360
361 ENLQLLRQRY YYATWKADGT RYMMLLTTDG CYIVDRSFRF RRVQMRFPFR HPTEGISDKV 420
421 HHFTLLDGEM IIDTLPDKQK QERRYLIYDM VAINGQSVVE VGIVIIYLCN Q |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
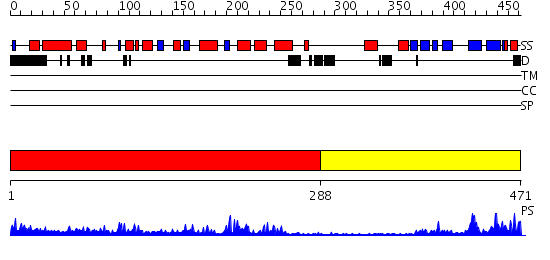
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..287] | 25.0 | CRYSTAL STRUCTURE OF THE HUMAN RNA guanylyltransferase and 5'- phosphatase | |
| 2 | View Details | [288..471] | 20.39794 | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)