













| General Information: |
|
| Name(s) found: |
BRM_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 2193 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] chromatin remodeling complex [IDA] |
| Biological Process: |
ATP-dependent chromatin remodeling
[NAS][TAS]
organ boundary specification between lateral organs and the meristem [IGI] regulation of gene expression, epigenetic [IMP] |
| Molecular Function: |
ATP binding
[ISS]
DNA binding [IDA][ISS] transcription regulator activity [ISS] helicase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSGGSGGGP ARNPAMGPAG RTASTSSAAS PSSSSSSVQQ QQQQQQQQQQ QQQLASRQQQ 60
61 QQHRNSDTNE NMFAYQPGGV QGMMGGGNFA SSPGSMQMPQ QSRNFFESPQ QQQQQQQQGS 120
121 STQEGQQNFN PMQQAYIQFA MQAQHQKAQQ QARMGMVGSS SVGKDQDARM GMLNMQDLNP 180
181 SSQPQASSSK PSGDQFARGE RQTESSSQQR NETKSHPQQQ VGTGQLMPGN MIRPMQAPQA 240
241 QQLVNNMGNN QLAFAQQWQA MQAWARERNI DLSHPANASQ MAHILQARMA AQQKAGEGNV 300
301 ASQSPSIPIS SQPASSSVVP GENSPHANSA SDISGQSGSA KARHALSTGS FASTSSPRMV 360
361 NPAMNPFSGQ GRENPMYPRH LVQPTNGMPS GNPLQTSANE TPVLDQNAST KKSLGPAEHL 420
421 QMQQPRQLNT PTPNLVAPSD TGPLSNSSLQ SGQGTQQAQQ RSGFTKQQLH VLKAQILAFR 480
481 RLKKGEGSLP PELLQAISPP PLELQTQRQI SPAIGKVQDR SSDKTGEDQA RSLECGKESQ 540
541 AAASSNGPIF SKEEDNVGDT EVALTTGHSQ LFQNLGKEAT STDVATKEEQ QTDVFPVKSD 600
601 QGADSSTQKN PRSDSTADKG KAVASDGSQS KVPPQANSPQ PPKDTASARK YYGPLFDFPF 660
661 FTRKLDSYGS ATANANNNLT LAYDIKDLIC EEGAEFLSKK RTDSLKKING LLAKNLERKR 720
721 IRPDLVLRLQ IEEKKLRLSD LQSRVREEVD RQQQEIMSMP DRPYRKFVRL CERQRLEMNR 780
781 QVLANQKAVR EKQLKTIFQW RKKLLEAHWA IRDARTARNR GVAKYHEKML REFSKRKDDG 840
841 RNKRMEALKN NDVERYREML LEQQTNMPGD AAERYAVLSS FLTQTEDYLH KLGGKITATK 900
901 NQQEVEEAAN AAAVAARLQG LSEEEVRAAA TCAREEVVIR NRFTEMNAPK ENSSVNKYYT 960
961 LAHAVNEVVV RQPSMLQAGT LRDYQLVGLQ WMLSLYNNKL NGILADEMGL GKTVQVMALI1020
1021 AYLMEFKGNY GPHLIIVPNA VLVNWKSELH TWLPSVSCIY YVGTKDQRSK LFSQEVCAMK1080
1081 FNVLVTTYEF IMYDRSKLSK VDWKYIIIDE AQRMKDRESV LARDLDRYRC QRRLLLTGTP1140
1141 LQNDLKELWS LLNLLLPDVF DNRKAFHDWF AQPFQKEGPA HNIEDDWLET EKKVIVIHRL1200
1201 HQILEPFMLR RRVEDVEGSL PAKVSVVLRC RMSAIQSAVY DWIKATGTLR VDPDDEKLRA1260
1261 QKNPIYQAKI YRTLNNRCME LRKACNHPLL NYPYFNDFSK DFLVRSCGKL WILDRILIKL1320
1321 QRTGHRVLLF STMTKLLDIL EEYLQWRRLV YRRIDGTTSL EDRESAIVDF NDPDTDCFIF1380
1381 LLSIRAAGRG LNLQTADTVV IYDPDPNPKN EEQAVARAHR IGQTREVKVI YMEAVVEKLS1440
1441 SHQKEDELRS GGSVDLEDDM AGKDRYIGSI EGLIRNNIQQ YKIDMADEVI NAGRFDQRTT1500
1501 HEERRMTLET LLHDEERYQE TVHDVPSLHE VNRMIARSEE EVELFDQMDE EFDWTEEMTN1560
1561 HEQVPKWLRA STREVNATVA DLSKKPSKNM LSSSNLIVQP GGPGGERKRG RPKSKKINYK1620
1621 EIEDDIAGYS EESSEERNID SGNEEEGDIR QFDDDELTGA LGDHQTNKGE FDGENPVCGY1680
1681 DYPPGSGSYK KNPPRDDAGS SGSSPESHRS KEMASPVSSQ KFGSLSALDT RPGSVSKRLL1740
1741 DDLEEGEIAA SGDSHIDLQR SGSWAHDRDE GDEEQVLQPT IKRKRSIRLR PRQTAERVDG1800
1801 SEMPAAQPLQ VDRSYRSKLR TVVDSHSSRQ DQSDSSSRLR SVPAKKVAST SKLHVSSPKS1860
1861 GRLNATQLTV EDNAEASRET WDGTSPISSS NAGARMSHII QKRCKIVISK LQRRIDKEGQ1920
1921 QIVPMLTNLW KRIQNGYAAG GVNNLLELRE IDHRVERLEY AGVMELASDV QLMLRGAMQF1980
1981 YGFSHEVRSE AKKVHNLFFD LLKMSFPDTD FREARNALSF SGSAPTLVST PTPRGAGISQ2040
2041 GKRQKLVNEP ETEPSSPQRS QQRENSRIRV QIPQKETKLG GTTSHTDESP ILAHPGELVI2100
2101 CKKKRKDREK SGPKTRTGGS SSPVSPPPAM IGRGLRSPVS GGVPRETRLA QQQRWPNQPT2160
2161 HPNNSGAAGD SVGWANPVKR LRTDSGKRRP SHL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
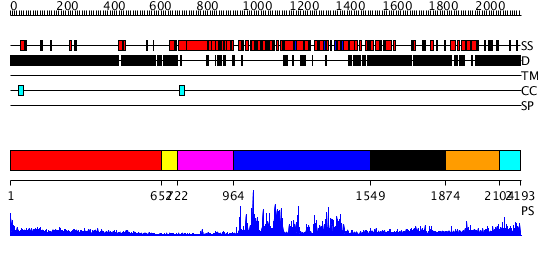
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..651] | 39.09691 | No description for 1y0fB was found. | |
| 2 | View Details | [652..721] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [722..963] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [964..1548] | 145.0 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 5 | View Details | [1549..1873] | 34.69897 | nucleosome recognition module of ISWI ATPase | |
| 6 | View Details | [1874..2103] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [2104..2193] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)