













| General Information: |
|
| Name(s) found: |
AGD4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 775 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
regulation of ARF GTPase activity
[IEA]
|
| Molecular Function: |
zinc ion binding
[IEA]
ARF GTPase activator activity [IEA] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATFINLEDS PMFQKQVCSL EGTADELKDR CQKLYKGVKK FMGTLGEASK GESAFAACLE 60
61 EFGGGPDDPI SLSIGGPVIS KFINALRELA SYKEFLCSQV EHVLLERLMN FISVDLQEAK 120
121 ESRHRFDKAA HSYDQSREKF VSLKKNTRGE IVAELEEDLE NSKSTFEKSR FNLVNSLMTI 180
181 EAKKKYEFLE SISAIMDAHL RYFKLGYDLL NQLEPFIHQI LTYAQQSKEQ SKIEQDRLAR 240
241 RIQEFRTQSE LDSQQLVANA ESSGANGNRV GGNIPYKNTE TSLTADKEVI KQGYLLKRSS 300
301 SLRTDWKRKF FVLDSHGSMY YYRTNGNKSM GSHHHYSGSS DHNTGVFGRF RARHNRSGSL 360
361 TEGSLGYNTI DLRTSLIKLD AEDMDLRLCF RIISPQKTYT LQAENGADRM DWVNKITKAI 420
421 GTLLNSHFLQ QSPVRYLDKD NSSSAPANAV VSGDQIRHND SRQNIGDDVS TILRGLPGNN 480
481 ACAECNAPEP DWASLNLGVL LCIQCSGVHR NLGVHISKVR SLSLDVKVWE PTILDLFRNL 540
541 GNVYCNSLWE GLLHLDDDCE DGSALSHASV SKPCPEDSFS VKEKYILGKY LEKALVIKDE 600
601 SEANLSAASR IWEAVQSRNI REIYRLIVTT GDVNIINTKF DDITDIDAYH HIDAAEKAVK 660
661 KRHDPTVCQR IKESNEPRSC LQGCSLLHVA CHIGDSVLLE LLLQFGADLN IRDYHGRTPL 720
721 HHCISSGNHK FAKILLRRGA RPSIEDDGGL SVLERAMEMG AITDEELFLL LAECA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
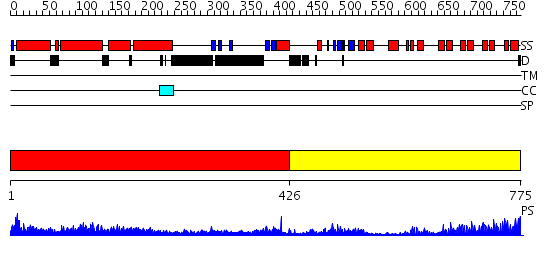
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..425] | 62.045757 | No description for 2elbA was found. | |
| 2 | View Details | [426..775] | 55.69897 | Ankyrin-R |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)