













| General Information: |
|
| Name(s) found: |
GLYC7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 598 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
L-serine metabolic process
[ISS]
response to cadmium ion [IEP] glycine metabolic process [ISS] |
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
catalytic activity [IEA] glycine hydroxymethyltransferase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLSRSQTNF QLGFGCSHAS MTPTPTPRAP IADDSINLQV DQSFRSLPTT FSPIPLQLLE 60
61 QKAEKTTTVD EPKKDGGGGG DQKEDEHFRI LGHHMCLKRQ RDCPLLLTQS KHPKRSSIGD 120
121 SDLESRRAAV RAWGDQPIHL ADPDIHELME KEKQRQVRGI ELIASENFVC RAVMEALGSH 180
181 LTNKYSEGMP GARYYTGNQY IDQIENLCIE RALTAFGLES DKWGVNVQPY SCTSANFAVY 240
241 TGLLLPGERI MGLDSPSGGH MSHGYCTPGG KKISAASIFF ESFPYKVNPQ TGYIDYDKLE 300
301 DKALDYRPKI LICGGSSYPR DWDFARVRQI ADKCGAVLMC DMAHISGLVA TKECSNPFDH 360
361 CDIVTSTTHK GLRGPRGGII FYRRGPKIRK QGHHSSHCDT STHYDLEEKI NFAVFPSLQG 420
421 GPHNNHIAAL AIALKQVATP EYKAYIQQMK KNAQALAAAL LRRKCRLVTG GTDNHLLLWD 480
481 LTPMGLTGKV YEKVCEMCHI TLNKTAIFGD NGTISPGGVR IGTPAMTTRG CIESDFETMA 540
541 DFLIKAAQIT SALQREHGKS HKEFVKSLCT NKDIAELRNR VEAFALQYEM PASLIRIE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
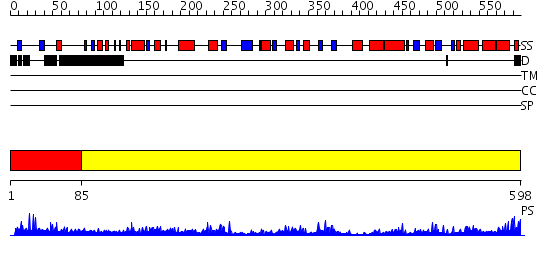
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [85..598] | 93.522879 | Human mitochondrial serine hydroxymethyltransferase 2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)