













| General Information: |
|
| Name(s) found: |
TCPG_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 555 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cellular protein metabolic process
[IEA]
protein folding [IEA] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] unfolded protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHAPVLVLSD SLKRESGSKV HHGNIQASKA VADIIRTTLG PRSMLKMLLD AGGGIVVTND 60
61 GNAILRELDV AHPAAKSMIE LSRTQDEEVG DGTTSVIVLA GEMLHVAEAF LEKNYHPTVI 120
121 CRAYIKALED SIAVLDKIAM SIDINDRSQV LGLVKSCIGT KFTSQFGDLI ADLAIDATTT 180
181 VGVDLGQGLR EVDIKKYIKV EKVPGGQFED SEVLKGVMFN KDVVAPGKMK RKIVNPRIIL 240
241 LDCPLEYKKG ENQTNAELVR EEDWEVLLKL EEEYIENICV QILKFKPDLV ITEKGLSDLA 300
301 CHYFSKAGVS AIRRLRKTDN NRIAKACGAV IVNRPDELQE SDIGTGAGLF EVKKIGDDFF 360
361 SFIVDCKEPK ACTVLLRGPS KDFINEVERN LQDAMSVARN IIKNPKLVPG GGATELTVSA 420
421 TLKQKSATIE GIEKWPYEAA AIAFEAIPRT LAQNCGVNVI RTMTALQGKH ANGENAWTGI 480
481 DGNTGAIADM KESKIWDSYN VKAQTFKTAI EAACMLLRID DIVSGIKKKQ APGSGPSKPT 540
541 IETEGDADNE QILPD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
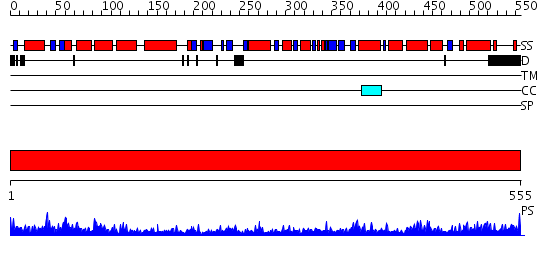
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..555] | 140.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)