













| General Information: |
|
| Name(s) found: |
gi|53791802
[NCBI NR]
gi|215695084 [NCBI NR] gi|115468554 [NCBI NR] gi|113595916 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 535 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALAFDEFGR PFIILREQEK KSRLRGLDAQ KANIAAGKAV ARILRTSLGP KGMDKMLQSP 60
61 DGDVTITNDG ATILEQMDVD NQIAKLMVEL SCSQDYEIGD GTTGVVVMAG SLLEQAEKLL 120
121 ERGIHPIRIA EGYELASRIA FDHLEHISHK FEFSATNIEP LVQTCMTTLS SKIVNRCKRT 180
181 LAEIAVKAVL AVADLERKDV NLDLIKVEGK VGGKLEDTEL VYGIIVDKDM SHPQMPKRIE 240
241 DAKIAILTCP FEPPKPKTKH KVDIDTVEKF QMLREQEQKY FDEMVQKCKD VGATLVICQW 300
301 GFDDEANHLL MHRNLPAVRW VGGVELELIA IATGGRIVPR FQELSPEKLG KAGIVREKSF 360
361 GTTKDRMLYI EQCANSRAVT IFIRGGNKMM IEETKRSLHD ALCVARNLIR NNSIVYGGGS 420
421 AEISCSVAVE AAADRYPGVE QYAIRSFADA LDAIPLALAE NSGLSPIDTL TAVKSQQVKE 480
481 SNPHCGIDCN DVGTNHMKEQ NVFETLIGKQ QQILLATQVV KMILKIDDVI SPSDY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
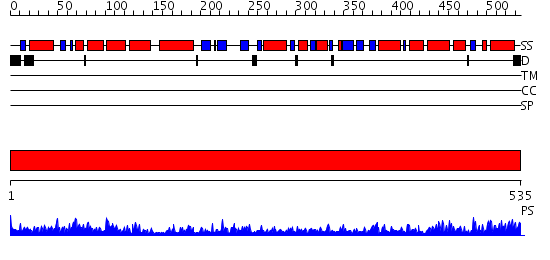
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..535] | 142.0 | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form of single mutant) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)