













| General Information: |
|
| Name(s) found: |
ORTH5_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
[NOT]DNA methylation on cytosine
[IMP]
protein ubiquitination [IDA] |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA]
zinc ion binding [ISS] protein binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIQTQLPCD GDGVCMRCQV NPPSEETLTC GTCVTPWHVS CLLPESLASS TGDWECPDCS 60
61 GVVVPSAAPG TGISGPESSG SVLVTAIRAI QADVTLTEAE KAKKRQRLMS GGGDDGVDDE 120
121 EKKKLEIFCS ICIQLPERPV TTPCGHNFCL KCFEKWAVGQ GKLTCMICRS KIPRHVAKNP 180
181 RINLALVSAI RLANVTKCSG EATAAKVHHI IRNQDRPDKA FTTERAVKTG KANAASGKFF 240
241 VTIPRDHFGP IPAANDVTRN QGVLVGESWE DRQECRQWGV HFPHVAGIAG QAAVGAQSVA 300
301 LSGGYDDDED HGEWFLYTGS GGRDLSGNKR VNKIQSSDQA FKNMNEALRL SCKMGYPVRV 360
361 VRSWKEKRSA YAPAEGVRYD GVYRIEKCWS NVGVQGLHKM CRYLFVRCDN EPAPWTSDEH 420
421 GDRPRPLPDV PELENATDLF VRKESPSWGF DEAEGRWKWM KSPPVSRMAL DTEERKKNKR 480
481 AKKGNNAMKA RLLKEFSCQI CRKVLSLPVT TPCAHNFCKA CLEAKFAGIT QLRDRSNGVR 540
541 KLRAKKNIMT CPCCTTDLSE FLQNPQVNRE MMEIIENFKK SEEEAEVAES SNISEEEGEE 600
601 ESEPPTKKIK MDKNSVGGTS LSA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
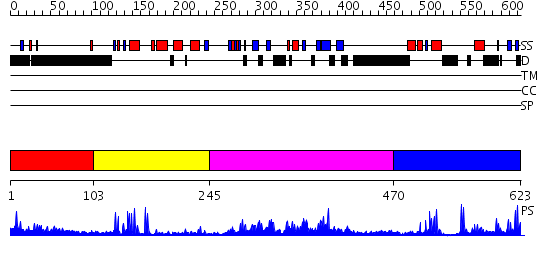
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 3.3 | No description for 2e6rA was found. | |
| 2 | View Details | [103..244] | 6.522879 | No description for 2cklB was found. | |
| 3 | View Details | [245..469] | 74.39794 | No description for PF02182.8 was found. No confident structure predictions are available. | |
| 4 | View Details | [470..623] | 17.0 | RAG1 DIMERIZATION DOMAIN |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)