













| General Information: |
|
| Name(s) found: |
gi|168267568
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | synthetic construct |
| Length: | 875 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHDAFEPVPI LEKLPLQIDC LAAWEEWLLV GTKQGHLLLY RIRKDVGCNR FEVTLEKSNK 60
61 NFSKKIQQIH VVSQFKILVS LLENNIYVHD LLTFQQITTV SKAKGASLFT CDLQHTETGE 120
121 EVLRMCVAVK KKLQLYFWKD REFHELQGDF SVPDVPKSMA WCENSICVGF KRDYYLIRVD 180
181 GKGSIKELFP TGKQLEPLVA PLADGKVAVG QDDLTVVLNE EGICTQKCAL NWTDIPVAME 240
241 HQPPYIIAVL PRYVEIRTFE PRLLVQSIEL QRPRFITSGG SNIIYVASNH FVWRLIPVPM 300
301 ATQIQQLLQD KQFELALQLA EMKDDSDSEK QQQIHHIKNL YAFNLFCQKR FDESMQVFAK 360
361 LGTDPTHVMG LYPDLLPTDY RKQLQYPNPL PVLSGAELEK AHLALIDYLT QKRSQLVKKL 420
421 NDSDHQSSTS PLMEGTPTIK SKKKLLQIID TTLLKCYLHT NVALVAPLLR LENNHCHIEE 480
481 SEHVLKKAHK YSELIILYEK KGLHEKALQV LVDQSKKANS PLKGHERTVQ YLQHLGTENL 540
541 HLIFSYSVWV LRDFPEDGLK IFTEDLPEVE SLPRDRVLGF LIENFKGLAI PYLEHIIHVW 600
601 EETGSRFHNC LIQLYCEKVQ GLMKEYLLSF PAGKTPVPAG EEEGELGEYR QKLLMFLEIS 660
661 SYYDPGRLIC DFPFDGLLEE RALLLGRMGK HEQALFIYVH ILKDTRMAEE YCHKHYDRNK 720
721 DGNKDVYLSL LRMYLSPPSI HCLGPIKLEL LEPKANLQAA LQVLELHHSK LDTTKALNLL 780
781 PANTQINDIR IFLEKVLEEN AQKKRFNQVL KNLLHAEFLR VQEERILHQQ VKCIITEEKV 840
841 CMVCKKKIGN SAFARYPNGV VVHYFCSKEV NPADT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
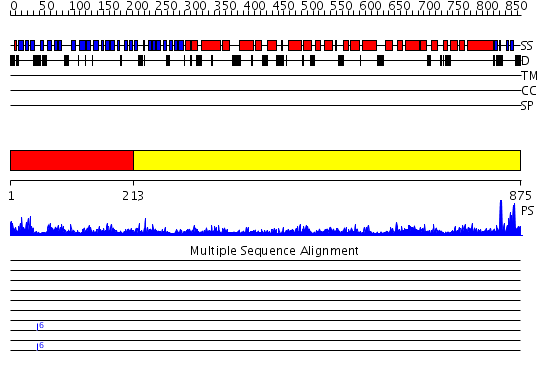
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..212] | 2.92 | Tup1, C-terminal domain | |
| 2 | View Details | [213..875] | 75.30103 | Clathrin D6 Coat |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)