













| General Information: |
|
| Name(s) found: |
gi|194902305
[NCBI NR]
gi|190652371 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Drosophila erecta |
| Length: | 487 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQPRYDDGL HGYGMDSGAA AAAMYDPHAG HRPPGLQGLP SHHSPHMTHA AAAAATVGMH 60
61 GYHSGAGGHG TPSHVSPVGN HLMGAIPEVH KRDKDAIYEH PLFPLLALIF EKCELATCTP 120
121 REPGVQGGDV CSSESFNEDI AMFSKQIRSQ KPYYTADPEV DSLMVQAIQV LRFHLLELEK 180
181 VHELCDNFCH RYISCLKGKM PIDLVIDERD TTKPPELGSA NGEGRSNADS TSHTDGASTP 240
241 DVRPPSSSLS YGGAMNDDAR SPGAGSTPGP LSQQPPALDT SDPDGKFLSS LNPSELTYDG 300
301 RWCRREWSSP ADARNADASR RLYSSVFLGS PDNFGTSASG DASNASIGSG EGTGEEDDDA 360
361 SGKKNQKKRG IFPKVATNIL RAWLFQHLTH PYPSEDQKKQ LAQDTGLTIL QVNNWFINAR 420
421 RRIVQPMIDQ SNRAVYTPHP GPSGYGHDAM GYMMDSQAHM MHRPPGDPGF HQGYPHYPPA 480
481 EYYGQHL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
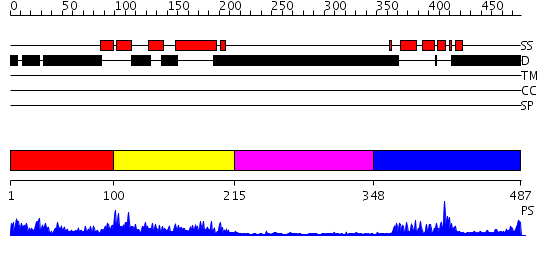
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | 1.053991 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [100..214] | 3.625969 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [215..347] | 4.550996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [348..487] | 11.0 | Solution structure of the homeobox domain of human homeobox protein PKNOX1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)