













| General Information: |
|
| Name(s) found: |
gi|221318794
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Bacillus subtilis subsp. subtilis str. JH642 |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGQEKHVFTI DWAGRTLTVE TGQLAKQANG AVMIRYGDTA VLSTATASKE PKPLDFFPLT 60
61 VNYEERLYAV GKIPGGFIKR EGRPSEKAVL ASRLIDRPIR PLFADGFRNE VQVISIVMSV 120
121 DQNCSSEMAA MFGSSLALSV SDIPFEGPIA GVTVGRIDDQ FIINPTVDQL EKSDINLVVA 180
181 GTKDAINMVE AGADEVPEEI MLEAIMFGHE EIKRLIAFQE EIVAAVGKEK SEIKLFEIDE 240
241 ELNEKVKALA EEDLLKAIQV HEKHAREDAI NEVKNAVVAK FEDEEHDEDT IKQVKQILSK 300
301 LVKNEVRRLI TEEKVRPDGR GVDQIRPLSS EVGLLPRTHG SGLFTRGQTQ ALSVCTLGAL 360
361 GDVQILDGLG VEESKRFMHH YNFPQFSVGE TGPMRGPGRR EIGHGALGER ALEPVIPSEK 420
421 DFPYTVRLVS EVLESNGSTS QASICASTLA MMDAGVPIKA PVAGIAMGLV KSGEHYTVLT 480
481 DIQGMEDALG DMDFKVAGTE KGVTALQMDI KIEGLSREIL EEALQQAKKG RMEILNSMLA 540
541 TLSESRKELS RYAPKILTMT INPDKIRDVI GPSGKQINKI IEETGVKIDI EQDGTIFISS 600
601 TDESGNQKAK KIIEDLVREV EVGQLYLGKV KRIEKFGAFV EIFSGKDGLV HISELALERV 660
661 GKVEDVVKIG DEILVKVTEI DKQGRVNLSR KAVLREEKEK EEQQS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
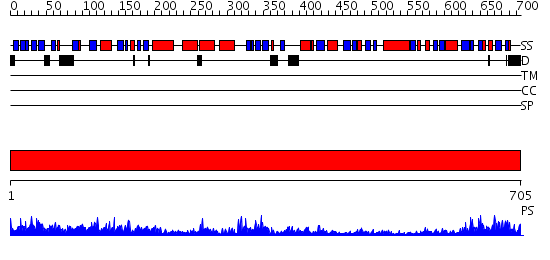
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..705] | 152.0 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)