













| General Information: |
|
| Name(s) found: |
gi|221317120
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Bacillus subtilis subsp. subtilis str. JH642 |
| Length: | 1177 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNIQTFIKE SDDFKSIING LHEGLKEQLL AGLSGSARSV FTSALANETN KPIFLITHNL 60
61 YQAQKVTDDL TSLLEDRSVL LYPVNELISS EIAVASPELR AQRLDVINRL TNGEAPIVVA 120
121 PVAAIRRMLP PVEVWKSSQM LIQVGHDIEP DQLASRLVEV GYERSDMVSA PGEFSIRGGI 180
181 IDIYPLTSEN PVRIELFDTE VDSIRSFNSD DQRSIETLTS INIGPAKELI IRPEEKARAM 240
241 EKIDSGLAAS LKKLKADKQK EILHANISHD KERLSEGQTD QELVKYLSYF YEKPASLLDY 300
301 TPDNTLLILD EVSRIHEMEE QLQKEEAEFI TNLLEEGKIL HDIRLSFSFQ KIVAEQKRPL 360
361 LYYSLFLRHV HHTSPQNIVN VSGRQMQSFH GQMNVLAGEM ERFKKSNFTV VFLGANKERT 420
421 QKLSSVLADY DIEAAMTDSK KALVQGQVYI MEGELQSGFE LPLMKLAVIT EEELFKNRVK 480
481 KKPRKQKLTN AERIKSYSEL QIGDYVVHIN HGIGKYLGIE TLEINGIHKD YLNIHYQGSD 540
541 KLYVPVEQID QVQKYVGSEG KEPKLYKLGG SEWKRVKKKV ETSVQDIADD LIKLYAEREA 600
601 SKGYAFSPDH EMQREFESAF PYQETEDQLR SIHEIKKDME RERPMDRLLC GDVGYGKTEV 660
661 AIRAAFKAIG DGKQVALLVP TTILAQQHYE TIKERFQDYP INIGLLSRFR TRKEANETIK 720
721 GLKNGTVDIV IGTHRLLSKD VVYKDLGLLI IDEEQRFGVT HKEKIKQIKA NVDVLTLTAT 780
781 PIPRTLHMSM LGVRDLSVIE TPPENRFPVQ TYVVEYNGAL VREAIERELA RGGQVYFLYN 840
841 RVEDIERKAD EISMLVPDAK VAYAHGKMTE NELETVMLSF LEGESDVLVS TTIIETGVDI 900
901 PNVNTLIVFD ADKMGLSQLY QLRGRVGRSN RVAYAYFTYR RDKVLTEVAE KRLQAIKEFT 960
961 ELGSGFKIAM RDLTIRGAGN LLGAQQHGFI DSVGFDLYSQ MLKEAIEERK GDTAKTEQFE1020
1021 TEIDVELDAY IPETYIQDGK QKIDMYKRFR SVATIEEKNE LQDEMIDRFG NYPKEVEYLF1080
1081 TVAEMKVYAR QERVELIKQD KDAVRLTISE EASAEIDGQK LFELGNQYGR QIGLGMEGKK1140
1141 LKISIQTKGR SADEWLDTVL GMLKGLKDVK KQTISST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
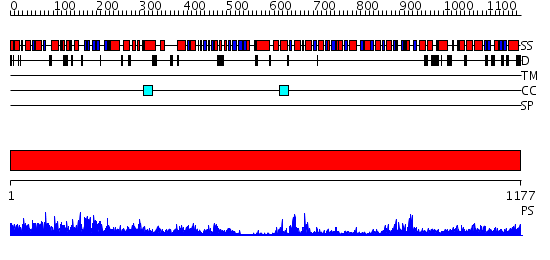
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1177] | 1000.0 | Crystal structure of Escherichia coli transcription-repair coupling factor |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)