













| General Information: |
|
| Name(s) found: |
gi|224502632
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Listeria monocytogenes FSL R2-561 |
| Length: | 544 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYSFKLPDI GEGIHEGEIV KWFVQPGDKI EEDESLFEVQ NDKSVEEITS PVSGTIKEIK 60
61 VAEGTVATVG QVLVTFDGVE GHEDDAEEES AAPKAESTES TPAPAQASGK GIFEFKLPDI 120
121 GEGIHEGEIV KWFIQPGDKV EEDQSIFEVQ NDKSVEEITS PVDGTVKDIL VSEGTVATVG 180
181 QVLVTFEGDF EGEASHESTP ESPAEDAALA NNDATSAPAT GGNGTPSSKK DPNGLVIAMP 240
241 SVRKYAREKG VNIAEVAGSG KNNRVVKADI DAFLNGEQPA AATTTAQTEE KAAAPKAEKA 300
301 AGKQPVASSD AYPETREKLT PTRRAIAKAM VNSKHTAPHV TLMDEIEVTA LMAHRKRFKE 360
361 VAAEKGIKLT FLPYMVKALV ATLRDFPVLN TTLDDATEEL VYKHYFNVGI AADTDHGLYV 420
421 PVIKNADKKS VFQISDEINE LAGKARDGKL TADEMRHGSA TISNIGSAGG QWFTPVINYP 480
481 EVAILGVGRI AQKPIVKDGE IVAAPVLALS LSFDHRVIDG ATAQKAMNNI KRLLNDPELL 540
541 LMEV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
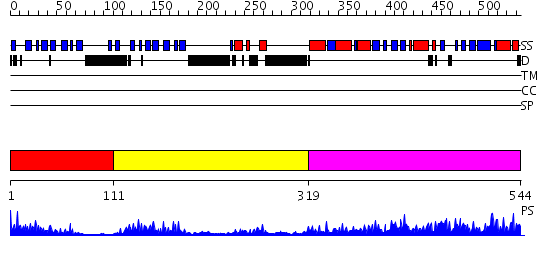
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] | 15.69897 | THREE-DIMENSIONAL STRUCTURE OF THE LIPOYL DOMAIN FROM BACILLUS STEAROTHERMOPHILUS PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX | |
| 2 | View Details | [111..318] | 23.154902 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 3 | View Details | [319..544] | 83.0 | Dihydrolipoamide acetyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)