













| General Information: |
|
| Name(s) found: |
gi|190405894
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 416 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTNTVPSSP PNQTPPAASG IATSHDHTKF NNPIRLPISI SLTINDTPNN NSNNNSVSNG 60
61 LGILPSRTAT SLVVANNGSA NGNVGATAAA AATVETNTAP AVNTTKSIRH FIYPPNQVNQ 120
121 TEFSLDIHLP PNTSLPERID QSTLKRRMDK HGLFSIRLTP FIDTSSTSVA NQGLFFDPII 180
181 RTAGAGSQII IGRYTERVRE AISKIPDQYH PVVFKSKVIS RTHGCFKVDD QGNWFLKDVK 240
241 SSSGTFLNHQ RLSSASTTSK DYLLHDGDII QLGMDFRGGT EEIYRCVKMK IELNKSWKLK 300
301 ANAFNKEALS RIKNLQKLTT GLEQEDCSIC LNKIKPCQAI FISPCAHSWH FHCVRRLVIM 360
361 NYPQFMCPNC RTNCDLETTL ESESESEFEN EDEDEPDIEM DIDMEINNNL GVRLVD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
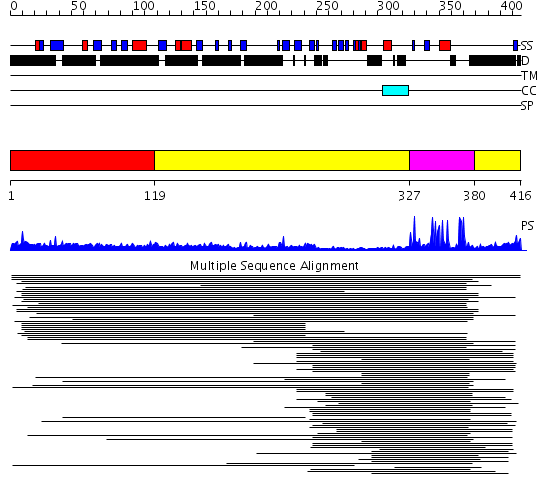
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [119..326] [380..416] |
6.0 | Cbl; N-terminal domain of cbl (N-cbl); CBL | |
| 3 | View Details | [327..379] | 6.0 | Cbl; N-terminal domain of cbl (N-cbl); CBL |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)