













| General Information: |
|
| Name(s) found: |
gi|220960168
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | synthetic construct |
| Length: | 873 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGSGGVANS SSTAAANNTA ATFQHIFEQL VQQGGGNHKL PPKQLEQLRH LLGNVRDAKN 60
61 LQMIVEKFKN LEQFHEHYAA HLANNNTVIS TEDSNDLVKD NARKYGSGGQ TLTPRHTIDA 120
121 ILGLKNRNGA ANGSGRNPET VSDGSVDPSL GDDDATDLRC GMTLTQLRSM DNHMASMLQQ 180
181 HAKNGGALPY GPPTPPGGQQ PQVPNATPLH HGQQMGGQAG HATHAGHGHP THHGHAPFGY 240
241 HNAFGFGQGG HGYGHPEEAA GNYLNSMHQM VEANQLQTGA NGANPPPAPL PPSSFGSHQQ 300
301 HLAALAAQAQ EQQNQHSKYA KSSPTGAGPP PPPGAYFMES QTAPVAPSQI NYDERSMSSA 360
361 SDLEEDDDDA AKLQLDVTSP PTPSPRGQLA AKRKSAGVFC DDNEPKLANG QLPGGNYGIR 420
421 PRSMEEVHHQ QQSHHHQQQQ QQQQQQQQLQ QQQGFQHDFR NSGNGNPNGN SNSGDHGERL 480
481 NADSDSLVNG SCASSEDLNQ TNSSEQGEKI TSGSDDEGQD DNCAKKKHRR NRTTFTTYQL 540
541 HELERAFEKS HYPDVYSREE LAMKVNLPEV RVQVWFQNRR AKWRRQEKSE SLRLGLTHFT 600
601 QLPHRLGCGA SGLPVDPWLS PPLLSALPGF LSHPQTVYPS YLTPPLSLAP GNLTMSSLAA 660
661 MGHHHAHNGP PPPHVGHGGH GQPQPPPPPP PHGVPHPHGS HHVVPLSHLS PHLSRMSPHA 720
721 TSLGSPHHGV TPLGTPLHSS LPPSSTATTV AVSSSQSSSS SASLECSGPD VCMSPQNLSI 780
781 GNADSNGDGR DLSSDLDAGS TSSNPGSSLD KCAASANIEL LDVGRDSPPP PPTPTGKGSS 840
841 TPPTDMRSNS IATLRIKAKE HLDNLNKGMV SIV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
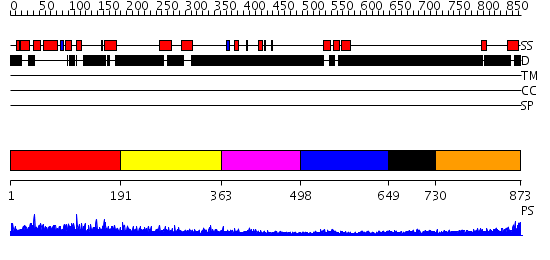
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [191..362] | 3.154902 | Sec24 | |
| 3 | View Details | [363..497] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [498..648] | 10.154902 | Solution structure of the homeobox domain of the human paired box protein Pax-6 | |
| 5 | View Details | [649..729] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [730..873] | 2.227996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)