













| General Information: |
|
| Name(s) found: |
Y4117_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1095 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
|
| Biological Process: |
response to ozone
[IEP]
defense response [ISS] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA] transmembrane receptor activity [IEA] protein binding [IEA] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASSSSNSWR YDVFPSFRGE DVRNNFLSHL LKEFESKGIV TFRDDHIKRS HTIGHELRAA 60
61 IRESKISVVL FSENYASSSW CLDELIEIMK CKEEQGLKVM PVFYKVDPSD IRKQTGKFGM 120
121 SFLETCCGKT EERQHNWRRA LTDAANILGD HPQNWDNEAY KITTISKDVL EKLNATPSRD 180
181 FNDLVGMEAH IAKMESLLCL ESQGVRIVGI WGPAGVGKTT IARALYNQYH ENFNLSIFME 240
241 NVRESYGEAG LDDYGLKLHL QQRFLSKLLD QKDLRVRHLG AIEERLKSQK VLIILDDVDN 300
301 IEQLKALAKE NQWFGNKSRI VVTTQNKQLL VSHDINHMYQ VAYPSKQEAL TIFCQHAFKQ 360
361 SSPSDDLKHL AIEFTTLAGH LPLALRVLGS FMRGKGKEEW EFSLPTLKSR LDGEVEKVLK 420
421 VGYDGLHDHE KDLFLHIACI FSGQHENYLK QMIIANNDTY VSFGLQVLAD KSLIQKFENG 480
481 RIEMHSLLRQ LGKEVVRKQS IYEPGKRQFL MNAKETCGVL SNNTGTGTVL GISLDMCEIK 540
541 EELYISEKTF EEMRNLVYLK FYMSSPIDDK MKVKLQLPEE GLSYLPQLRL LHWDAYPLEF 600
601 FPSSFRPECL VELNMSHSKL KKLWSGVQPL RNLRTMNLNS SRNLEILPNL MEATKLNRLD 660
661 LGWCESLVEL PSSIKNLQHL ILLEMSCCKK LEIIPTNINL PSLEVLHFRY CTRLQTFPEI 720
721 STNIRLLNLI GTAITEVPPS VKYWSKIDEI CMERAKVKRL VHVPYVLEKL CLRENKELET 780
781 IPRYLKYLPR LQMIDISYCI NIISLPKLPG SVSALTAVNC ESLQILHGHF RNKSIHLNFI 840
841 NCLKLGQRAQ EKIHRSVYIH QSSYIADVLP GEHVPAYFSY RSTGSSIMIH SNKVDLSKFN 900
901 RFKVCLVLGA GKRFEGCDIK FYKQFFCKPR EYYVPKHLDS PLLKSDHLCM CEFELMPPHP 960
961 PTEWELLHPN EFLEVSFESR GGLYKCEVKE CGLQFLEPHE TSEFRYLSPH LYLGGSWIGN1020
1021 SSSSIEEIIH VDQEESSSDS EEIIYADQEE SSSGIEEIIH AEREGTNRRK SVMRWIKVGA1080
1081 RKMGLSLECL KPWTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
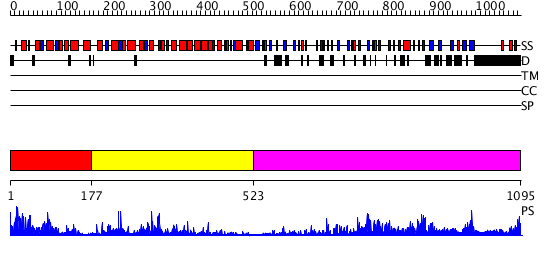
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 63.522879 | No description for PF01582.12 was found. No confident structure predictions are available. | |
| 2 | View Details | [177..522] | 13.69897 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [523..1095] | 21.39794 | No description for 3cigA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)