













| General Information: |
|
| Name(s) found: |
ANM15_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
positive regulation of vernalization response
[IMP]
regulation of flower development [IMP] |
| Molecular Function: |
protein methyltransferase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLGERGGWE RTESRYCGVE TDFSNDVTHL LNFNISTGGF DYVLAPLVDP SYRPSLVEGN 60
61 GVDTQVLPVC GSDLVLSPSQ WSSHVVGKIS SWIDLDSEDE VLRMDSETTL KQEIAWATHL 120
121 SLQACLLPTP KGKSCANYAR CVNQILQGLT TLQLWLRVPL VKSEGDSMDD TSEGLNDSWE 180
181 LWNSFRLLCE HDSKLSVALD VLSTLPSETS LGRWMGESVR AAILSTDAFL TNARGYPCLS 240
241 KRHQKLIAGF FDHAAQVVIC GKPVHNLQKP LDSSSEGTEK NPLRIYLDYV AYLFQKMESL 300
301 SEQERIELGY RDFLQAPLQP LMDNLEAQTY ETFERDSVKY IQYQRAVEKA LVDRVPDEKA 360
361 SELTTVLMVV GAGRGPLVRA SLQAAEETDR KLKVYAVEKN PNAVVTLHNL VKMEGWEDVV 420
421 TIISCDMRFW NAPEQADILV SELLGSFGDN ELSPECLDGA QRFLKPDGIS IPSSYTSFIQ 480
481 PITASKLYND VKAHKDLAHF ETAYVVKLHS VAKLAPSQSV FTFTHPNFST KVNNQRYKKL 540
541 QFSLPSDAGS ALVHGFAGYF DSVLYKDVHL GIEPTTATPN MFSWFPIFFP LRKPVEVHPD 600
601 TPLEVHFWRC CGSSKVWYEW SVSSPTPSPM HNTNGRSYWV GL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
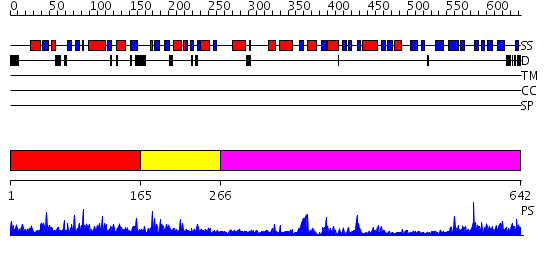
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | 1.048976 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [165..265] | 18.154902 | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | |
| 3 | View Details | [266..642] | 53.69897 | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)