













| General Information: |
|
| Name(s) found: |
PUB3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 760 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IEA]
|
| Biological Process: |
protein ubiquitination
[IEA]
|
| Molecular Function: |
ubiquitin-protein ligase activity
[IEA]
binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPVPVRCLL NSISRYLHLV ACQTIRFNPI QTCIGNMVLL LKLLKPLLDE VVDCKIPSDD 60
61 CLYKGCEDLD SVVNQAREFL EDWSPKLSKL FGVFQCEVLL GKVQTCSLEI SRILLQLSQS 120
121 SPVTSSVQSV ERCVQETESF KQEGTLMELM ENALRNQKDD ITSLDNNHLE SIIQMLGLIS 180
181 NQDLLKESIT VEKERIRSQA SKSEEDMEQT EQLIELVLCI REHMLKTEFL EVAKGISIPP 240
241 YFRCPLSTEL MLDPVIVASG QTFDRTSIKK WLDNGLAVCP RTRQVLTHQE LIPNYTVKAM 300
301 IASWLEANRI NLATNSCHQY DGGDASSMAN NMGSQDFNRT ESFRFSLRSS SLTSRSSLET 360
361 GNGFEKLKIN VSASLCGESQ SKDLEIFELL SPGQSYTHSR SESVCSVVSS VDYVPSVTHE 420
421 TESILGNHQS SSEMSPKKNL ESSNNVNHEH SAAKTYECSV HDLDDSGTMT TSHTIKLVED 480
481 LKSGSNKVKT AAAAEIRHLT INSIENRVHI GRCGAITPLL SLLYSEEKLT QEHAVTALLN 540
541 LSISELNKAM IVEVGAIEPL VHVLNTGNDR AKENSAASLF SLSVLQVNRE RIGQSNAAIQ 600
601 ALVNLLGKGT FRGKKDAASA LFNLSITHDN KARIVQAKAV KYLVELLDPD LEMVDKAVAL 660
661 LANLSAVGEG RQAIVREGGI PLLVETVDLG SQRGKENAAS VLLQLCLNSP KFCTLVLQEG 720
721 AIPPLVALSQ SGTQRAKEKA QQLLSHFRNQ RDARMKKGRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
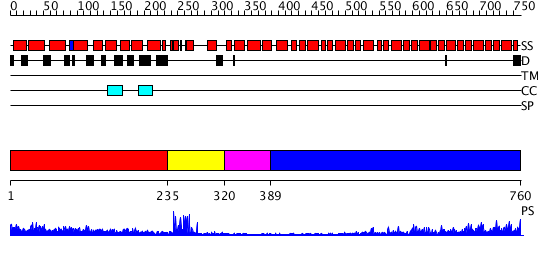
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..234] | 5.177993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [235..319] | 11.69897 | NMR solution structure of the U box domain from AtPUB14, an armadillo repeat containing protein from Arabidopsis thaliana | |
| 3 | View Details | [320..388] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [389..760] | 51.522879 | Importin alpha |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)