













| General Information: |
|
| Name(s) found: |
CKI1 /
YLR133W
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 582 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[TAS]
|
| Biological Process: |
phosphatidylcholine biosynthetic process
[TAS]
|
| Molecular Function: |
choline kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQESRPGSV RSYSVGYQAR SRSSSQRRHS LTRQRSSQRL IRTISIESDV SNITDDDDLR 60
61 AVNEGVAGVQ LDVSETANKG PRRASATDVT DSLGSTSSEY IEIPFVKETL DASLPSDYLK 120
121 QDILNLIQSL KISKWYNNKK IQPVAQDMNL VKISGAMTNA IFKVEYPKLP SLLLRIYGPN 180
181 IDNIIDREYE LQILARLSLK NIGPSLYGCF VNGRFEQFLE NSKTLTKDDI RNWKNSQRIA 240
241 RRMKELHVGV PLLSSERKNG SACWQKINQW LRTIEKVDQW VGDPKNIENS LLCENWSKFM 300
301 DIVDRYHKWL ISQEQGIEQV NKNLIFCHND AQYGNLLFTA PVMNTPSLYT APSSTSLTSQ 360
361 SSSLFPSSSN VIVDDIINPP KQEQSQDSKL VVIDFEYAGA NPAAYDLANH LSEWMYDYNN 420
421 AKAPHQCHAD RYPDKEQVLN FLYSYVSHLR GGAKEPIDEE VQRLYKSIIQ WRPTVQLFWS 480
481 LWAILQSGKL EKKEASTAIT REEIGPNGKK YIIKTEPESP EEDFVENDDE PEAGVSIDTF 540
541 DYMAYGRDKI AVFWGDLIGL GIITEEECKN FSSFKFLDTS YL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CKI1 NAP1 |
| View Details | Krogan NJ, et al. (2006) |

|
ABF2 ARG82 CKI1 CMR1 HHF1, HHF2 HTA1 HTA2 HTB1 HTB2 NUP188 PPH3 PSY2 PSY4 RRD1 SLX5 SLX8 SPT5 STP1 TOP2 |
| View Details | Ho Y, et al. (2002) |

|
CDC25 CKI1 GCR2 GIN4 HOM3 IRA1 IRA2 MYO2 MYO4 NAP1 PMD1 PRS2 PRS3 PRS5 RIM11 TPS1 TSL1 YDR170W-A YER138C YER160C YJR027W YJR028W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
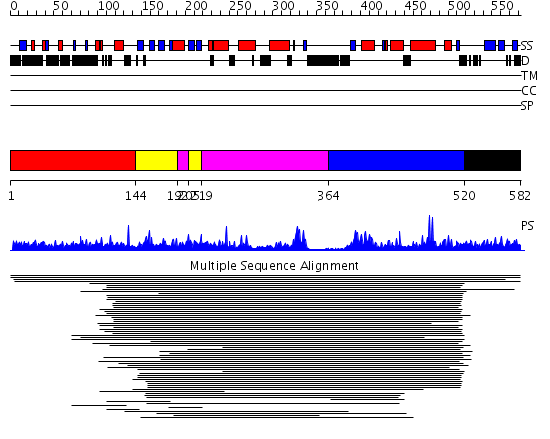
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [144..191] [205..218] |
12.95 | Type IIIa 3',5"-aminoglycoside phosphotransferase | |
| 3 | View Details | [192..204] [219..363] |
12.95 | Type IIIa 3',5"-aminoglycoside phosphotransferase | |
| 4 | View Details | [364..519] | 1.049982 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [520..582] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)