













| General Information: |
|
| Name(s) found: |
YKR017C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 551 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSGTENDQF YSFDESDSSS IELYESHNTS EFTIHGLVFP KLISVTSQDS EFDINEDEDG 60
61 VDTIYEGMLD APLTKNNKRI LCEGSVPNLS YECLTTKGIF ERMLQRVDHL QPIFAIPSAD 120
121 ILILLQHYDW NEERLLEVWT EKMDELLVEL GLSTTANIKK DNDYNSHFRE VEFKNDFTCI 180
181 ICCDKKDTET FALECGHEYC INCYRHYIKD KLHEGNIITC MDCSLALKNE DIDKVMGHPS 240
241 SSKLMDSSIK SFVQKHNRNY KWCPFADCKS IVHLRDTSSL PEYTRLHYSP FVKCNSFHRF 300
301 CFNCGFEVHS PADCKITTAW VKKARKESEI LNWVLSHTKE CPKCSVNIEK NGGCNHMVCS 360
361 SCKYEFCWIC EGPWAPHGKN FFQCTMYKNN EDNKSKNPQD ANKTLKKYTF YYRLFNEHEV 420
421 SAKLDWNLGQ TLGTKVHALQ ERIGISWIDG QFLSESLKVL NEGRTVLKWS FAVAYYSDAS 480
481 HNLTKIFVDN QMLLANAVES LSELLQIKTP EVIMKRRPEF YNKAGYVENR TTALMECGRE 540
541 LLCKGICKAA E |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
ETP1 MOH1 NHA1 YKR017C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
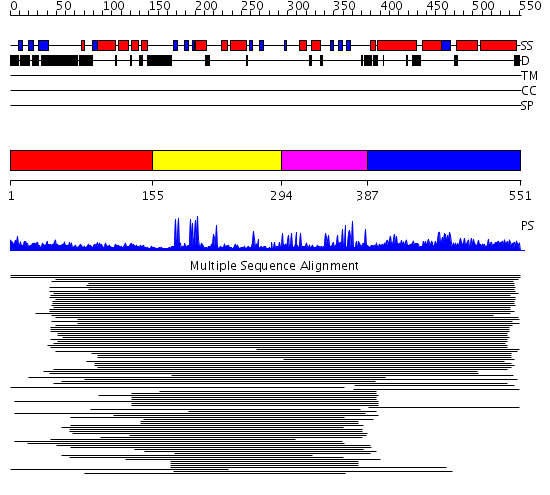
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..154] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [155..293] | 2.045757 | V(D)J recombination activating protein 1 (RAG1), dimerization domain | |
| 3 | View Details | [294..386] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [387..551] | 3.105962 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [392..551] | 7.15898 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)