













| General Information: |
|
| Name(s) found: |
UBX7 /
YBR273C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 436 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA endoplasmic reticulum [IDA |
| Biological Process: |
sporulation resulting in formation of a cellular spore
[IMP ubiquitin-dependent protein catabolic process [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLEALFRDSV EEAINDSIKE GVVLAVYNTA RDDQWLKSWF KGDDVSLDTL AEHSIWLRLV 60
61 KDTEQFQLFE QVFPNVVVPS IYLIRAGKIE LIIQGEDDRH WEKLLACIGI KDKKAGESSS 120
121 RETNPGLARE EKSSRDVHRK NARERIAETT LEIQRREQLK QRKLAEEERE RIIRLVRADR 180
181 AERKALDETH HRTLDDDKPL DVHDYIKDAQ KLHSSKCVLQ IRMTDGKTLK HEFNSSETLN 240
241 DVRKWVDVNR TDGDCPYSFH RGIPRVTFKD SDELKTLETL ELTPRSALLL KPLETQNSGL 300
301 SVTGMEGPSL LGRLYKGFST WWHNDKDPEV TSQREETSKP NRHEVRSSTP LSGAASSSCF 360
361 QYNNVREPVQ SSAHASPMLT PSGTRYPSET NLTTSRSVSP NVFQFVNNDH QEDPEDPTTF 420
421 NGNNVHLEKK KDEDKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
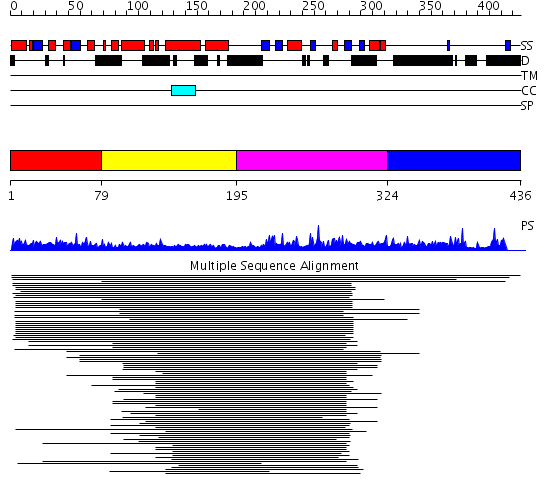
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [79..194] | 2.097993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [195..323] | 17.154902 | p47 | |
| 4 | View Details | [324..436] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)