













| General Information: |
|
| Name(s) found: |
FBpp0302532
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 562 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
polytene chromosome [IDA] DNA-directed RNA polymerase II, holoenzyme [ISS] Ada2/Gcn5/Ada3 transcription activator complex [NAS] H4/H2A histone acetyltransferase complex [IDA] DNA-directed RNA polymerase II, core complex [IDA][ISS] histone acetyltransferase complex [IDA] |
| Biological Process: |
mRNA transcription from RNA polymerase II promoter
[IDA]
cell proliferation [IDA] histone H3 acetylation [IDA] transcription [IEA] regulation of histone acetylation [IDA] transcription from RNA polymerase II promoter [IDA][ISS] |
| Molecular Function: |
nucleotide binding
[IEA]
histone acetyltransferase activity [NAS] DNA binding [IEA] zinc ion binding [IEA] DNA-directed RNA polymerase activity [IDA][ISS] protein binding [IPI] transcription regulator activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFMNPVDMV DEDAADLQFP KVALLHIFTT LNCLFAVEPL GPQVKRFIHA HQNEHTRNSM 60
61 VLYNPHTLHR YLEEMEEKTR DQNSSVPSAT KDANRCATCR CSLTEPYIKC SECLDTLLCL 120
121 QCFSRGKEAF SHRNNHAYII VRDNIQVFAD EPHWTARDER ILLKTLRTHG YGNWEAVSQA 180
181 LDQRHEPAEV RRHYHDCYFG GIFERLLNLK HARDSYVPER MPYVFKMRSL DPPRHDDIAS 240
241 MQFRLSAGYR CARGDFDTPY DTSAESLLSI MVDHRGRDDD NEASESEFER EVTEELQLGL 300
301 VRAYNNRLRE RQRRYKIMRQ HGLIMPNRTV SWISKYVHAF GSDASCMRFL GFMQICPDPI 360
361 KFDMLLESLR YYRELHSQLH KLYDLREHGV RTLSGAKLYA RLSKERQQAQ RDYSRLKQTD 420
421 AFDWQQLVQH YESNRSGDPG PLAINSKLYV MNTRRKASPI EIGGGKHFTH CLTPTEYNFS 480
481 LIPDLPGYSK LDDGERKLCS VARLVPQSYL DYKNQLVTEQ AKLGYLRLAD ARRLIKIDVN 540
541 KTRQIYDFLL EHGHISRPPS YG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
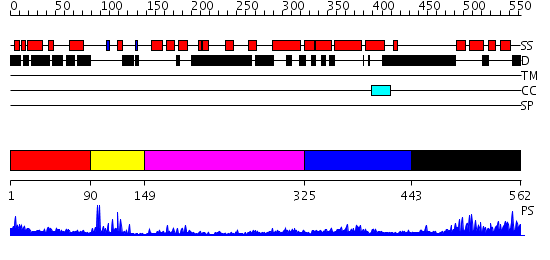
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 9.30103 | Dystrophin | |
| 2 | View Details | [90..148] | 7.30103 | No description for 2e5rA was found. | |
| 3 | View Details | [149..324] | 2.84 | CHICKEN B-MYB DNA BINDING DOMAIN, REPEAT 2 AND REPEAT3, NMR, 32 STRUCTURES | |
| 4 | View Details | [325..442] | 1.166992 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [443..562] | 26.0 | Solution structure of SWIRM domain of mouse transcriptional adaptor 2-like |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)