













| General Information: |
|
| Name(s) found: |
RECN_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 553 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLAQLTISNF AIVRELEIDF HSGMTVITGE TGAGKSIAID ALGLCLGGRA EADMVRTGAA 60
61 RADLCARFSL KDTPAALRWL EENQLEDGHE CLLRRVISSD GRSRGFINGT AVPLSQLREL 120
121 GQLLIQIHGQ HAHQLLTKPE HQKFLLDGYA NETSLLQEMT ARYQLWHQSC RDLAHHQQLS 180
181 QERAARAELL QYQLKELNEF NPQPGEFEQI DEEYKRLANS GQLLTTSQNA LALMADGEDA 240
241 NLQSQLYTAK QLVSELIGMD SKLSGVLDML EEATIQIAEA SDELRHYCDR LDLDPNRLFE 300
301 LEQRISKQIS LARKHHVSPE ALPQYYQSLL EEQQQLDDQA DSQETLALAV TKHHQQALEI 360
361 ARALHQQRQQ YAEELAQLIT DSMHALSMPH GQFTIDVKFD EHHLGADGAD RIEFRVTTNP 420
421 GQPMQPIAKV ASGGELSRIA LAIQVITARK METPALIFDE VDVGISGPTA AVVGKLLRQL 480
481 GESTQVMCVT HLPQVAGCGH QHYFVSKETD GAMTETHMQS LNKKARLQEL ARLLGGSEVT 540
541 RNTLANAKEL LAA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
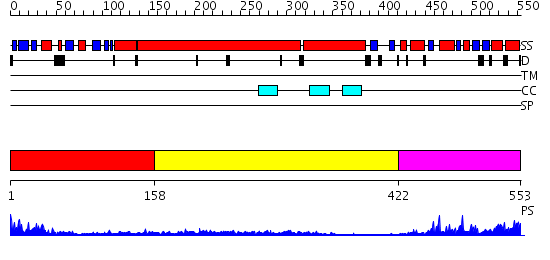
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 32.69897 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. | |
| 2 | View Details | [158..421] | 4.045757 | No description for 2i1jA was found. | |
| 3 | View Details | [422..553] | 3.69897 | No description for 2iw3A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)